| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
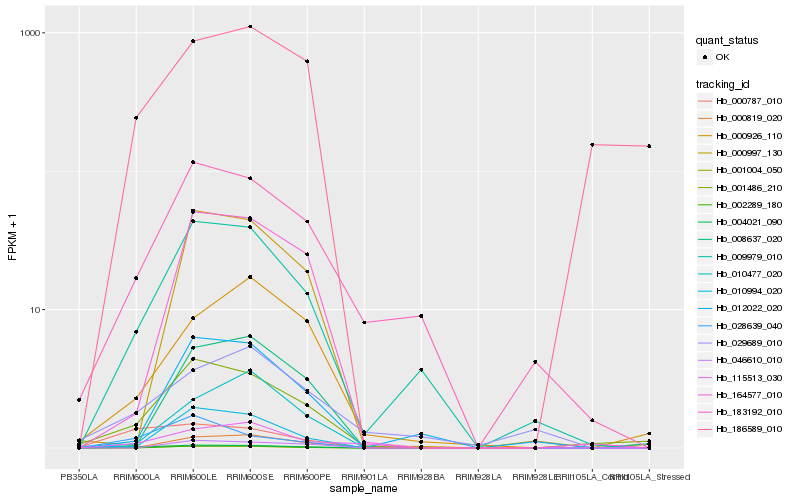
Hb_004021_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_09233 [Jatropha curcas] |
| 2 |
Hb_009979_010 |
0.1674395582 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_001486_210 |
0.1675817644 |
- |
- |
- |
| 4 |
Hb_000787_010 |
0.1753762078 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 5 |
Hb_028639_040 |
0.2049565479 |
- |
- |
PREDICTED: uncharacterized protein LOC105761689 [Gossypium raimondii] |
| 6 |
Hb_183192_010 |
0.2130860254 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 7 |
Hb_115513_030 |
0.2226356096 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 8 |
Hb_001004_050 |
0.223280389 |
- |
- |
PREDICTED: uncharacterized protein LOC103335231 [Prunus mume] |
| 9 |
Hb_164577_010 |
0.2244252064 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 10 |
Hb_000926_110 |
0.2383975369 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 11 |
Hb_000819_020 |
0.2424377666 |
- |
- |
hypothetical protein PRUPE_ppa020290mg, partial [Prunus persica] |
| 12 |
Hb_010994_020 |
0.2425211399 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_012022_020 |
0.2436403799 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 14 |
Hb_186589_010 |
0.2477157321 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Nelumbo nucifera] |
| 15 |
Hb_008637_020 |
0.2484670652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010477_020 |
0.2492469174 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 17 |
Hb_000997_130 |
0.2531436871 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_029689_010 |
0.257526418 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 19 |
Hb_002289_180 |
0.2591210556 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Nelumbo nucifera] |
| 20 |
Hb_046610_010 |
0.2594779729 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |