| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
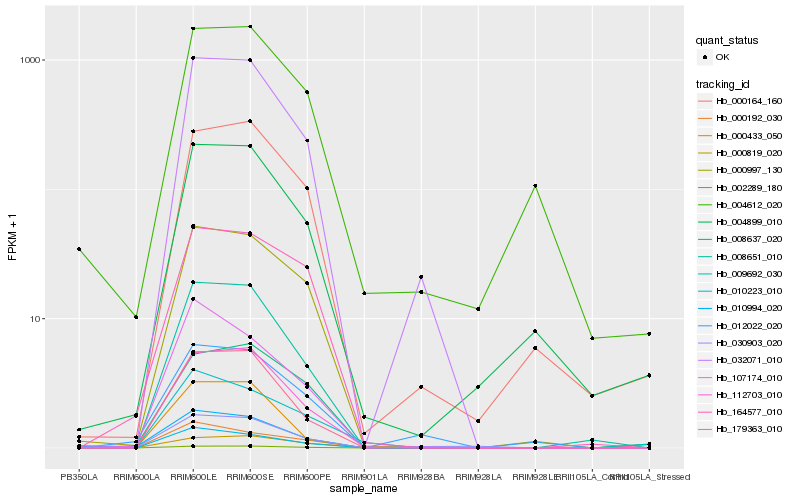
Hb_010994_020 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_008651_010 |
0.1215295959 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_010223_010 |
0.1244680971 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 4 |
Hb_107174_010 |
0.1318741553 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_030903_020 |
0.1330982951 |
- |
- |
PREDICTED: putative 12-oxophytodienoate reductase 11 [Jatropha curcas] |
| 6 |
Hb_000997_130 |
0.1331928828 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_179363_010 |
0.1347505015 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_032071_010 |
0.1389514405 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 9 |
Hb_112703_010 |
0.1394546722 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Prunus mume] |
| 10 |
Hb_002289_180 |
0.1408836957 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Nelumbo nucifera] |
| 11 |
Hb_004899_010 |
0.1577314824 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 12 |
Hb_164577_010 |
0.1624487035 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 13 |
Hb_000433_050 |
0.1625453379 |
- |
- |
PREDICTED: cell number regulator 1 isoform X1 [Vitis vinifera] |
| 14 |
Hb_012022_020 |
0.1627838842 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 15 |
Hb_009692_030 |
0.1633918642 |
- |
- |
WRKY DNA-binding protein 70, putative isoform 2, partial [Theobroma cacao] |
| 16 |
Hb_004612_020 |
0.166331346 |
- |
- |
PREDICTED: tetraspanin-8-like [Populus euphratica] |
| 17 |
Hb_000192_030 |
0.1705454874 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 18 |
Hb_000164_160 |
0.1749238233 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_008637_020 |
0.1800074492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000819_020 |
0.1811785117 |
- |
- |
hypothetical protein PRUPE_ppa020290mg, partial [Prunus persica] |