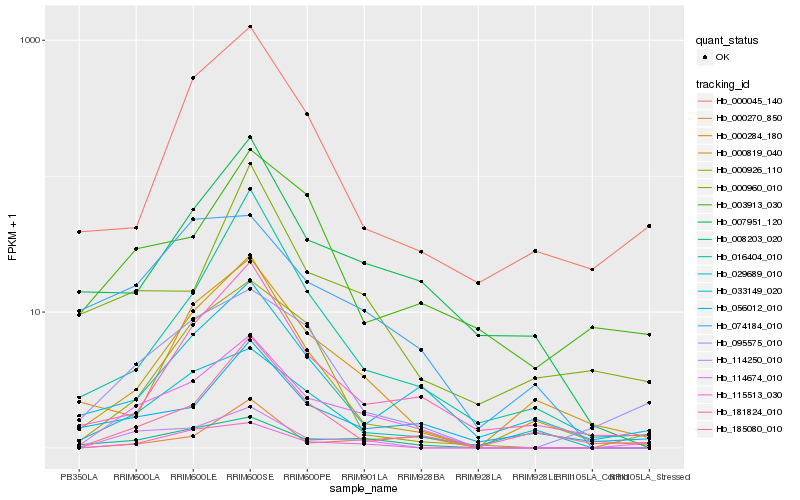
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114674_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 2 |
Hb_008203_020 |
0.1559589405 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 3 |
Hb_114250_010 |
0.1757973483 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_115513_030 |
0.1947649586 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 5 |
Hb_095575_010 |
0.2000600175 |
- |
- |
hypothetical protein JCGZ_19529 [Jatropha curcas] |
| 6 |
Hb_029689_010 |
0.2015024649 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 7 |
Hb_181824_010 |
0.2019034372 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_007951_120 |
0.2072812085 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 9 |
Hb_000819_040 |
0.2096462045 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_033149_020 |
0.2106543025 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 11 |
Hb_016404_010 |
0.2256367313 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_000926_110 |
0.2277555785 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 13 |
Hb_185080_010 |
0.2293435365 |
- |
- |
- |
| 14 |
Hb_056012_010 |
0.2320095299 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 15 |
Hb_000045_140 |
0.2337459619 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 16 |
Hb_000270_850 |
0.2348526368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000284_180 |
0.2362052839 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 18 |
Hb_074184_010 |
0.2413629568 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 19 |
Hb_000960_010 |
0.2431012945 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 20 |
Hb_003913_030 |
0.2436745687 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |