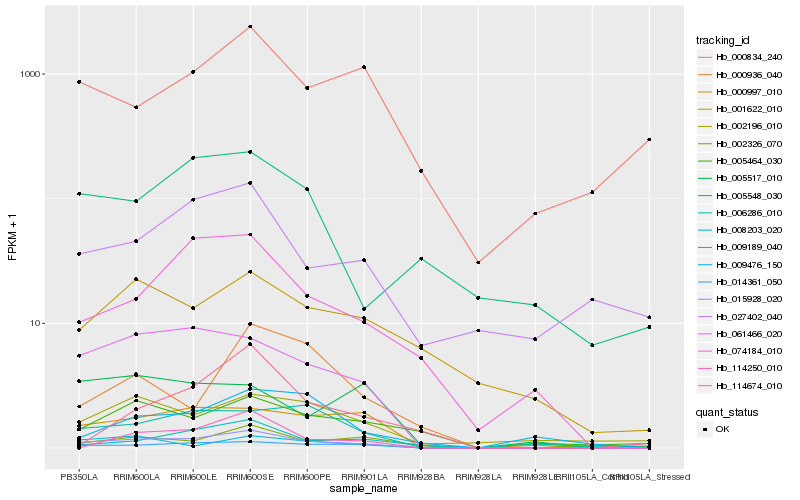
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015928_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 2 |
Hb_014361_050 |
0.1454558827 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 3 |
Hb_061466_020 |
0.1632067864 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 4 |
Hb_000997_010 |
0.1879387789 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 5 |
Hb_008203_020 |
0.213455538 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 6 |
Hb_074184_010 |
0.2169441627 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 7 |
Hb_006286_010 |
0.2296957326 |
- |
- |
hypothetical protein PRUPE_ppa017148mg [Prunus persica] |
| 8 |
Hb_009189_040 |
0.2402830758 |
- |
- |
PREDICTED: uncharacterized protein LOC104446949 [Eucalyptus grandis] |
| 9 |
Hb_114250_010 |
0.246185273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002196_010 |
0.2527924268 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: proline-rich receptor-like protein kinase PERK1 [Jatropha curcas] |
| 11 |
Hb_001622_010 |
0.2550315539 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_005464_030 |
0.2556007347 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_002326_070 |
0.2560662701 |
- |
- |
ent-kaurene oxidase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005517_010 |
0.2618130644 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000936_040 |
0.2620121522 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 16 |
Hb_027402_040 |
0.26358841 |
- |
- |
Thioredoxin II, putative [Ricinus communis] |
| 17 |
Hb_009476_150 |
0.2659600341 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 18 |
Hb_000834_240 |
0.2697811744 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 19 |
Hb_005548_030 |
0.2704415792 |
- |
- |
hypothetical protein JCGZ_05606 [Jatropha curcas] |
| 20 |
Hb_114674_010 |
0.2768925153 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |