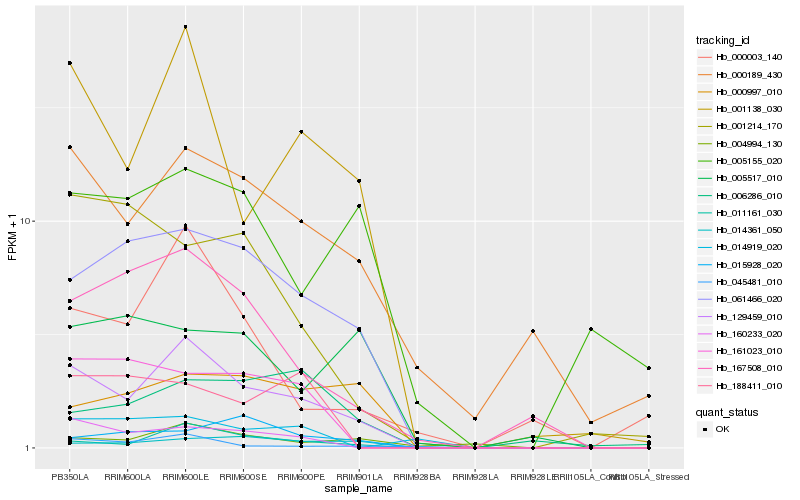
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129459_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 2 |
Hb_001138_030 |
0.1563224935 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 3 |
Hb_011161_030 |
0.1651923169 |
- |
- |
PREDICTED: uncharacterized protein LOC105638906 [Jatropha curcas] |
| 4 |
Hb_061466_020 |
0.1799224649 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 5 |
Hb_000189_430 |
0.1982096849 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_167508_010 |
0.2152526594 |
- |
- |
hypothetical protein POPTR_0001s27330g [Populus trichocarpa] |
| 7 |
Hb_160233_020 |
0.2229829034 |
- |
- |
- |
| 8 |
Hb_045481_010 |
0.2267985659 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000003_140 |
0.2375698003 |
- |
- |
hypothetical protein glysoja_047034, partial [Glycine soja] |
| 10 |
Hb_004994_130 |
0.2418966647 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |
| 11 |
Hb_014361_050 |
0.2463636725 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000997_010 |
0.2542499273 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 13 |
Hb_161023_010 |
0.2631347704 |
- |
- |
PREDICTED: uncharacterized protein LOC105639929 [Jatropha curcas] |
| 14 |
Hb_005155_020 |
0.2687949611 |
- |
- |
- |
| 15 |
Hb_005517_010 |
0.2739248658 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_006286_010 |
0.2759335023 |
- |
- |
hypothetical protein PRUPE_ppa017148mg [Prunus persica] |
| 17 |
Hb_001214_170 |
0.2761324062 |
- |
- |
pyruvate kinase family protein [Medicago truncatula] |
| 18 |
Hb_188411_010 |
0.281912898 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 [Jatropha curcas] |
| 19 |
Hb_015928_020 |
0.2830849743 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 20 |
Hb_014919_020 |
0.2861092283 |
- |
- |
- |