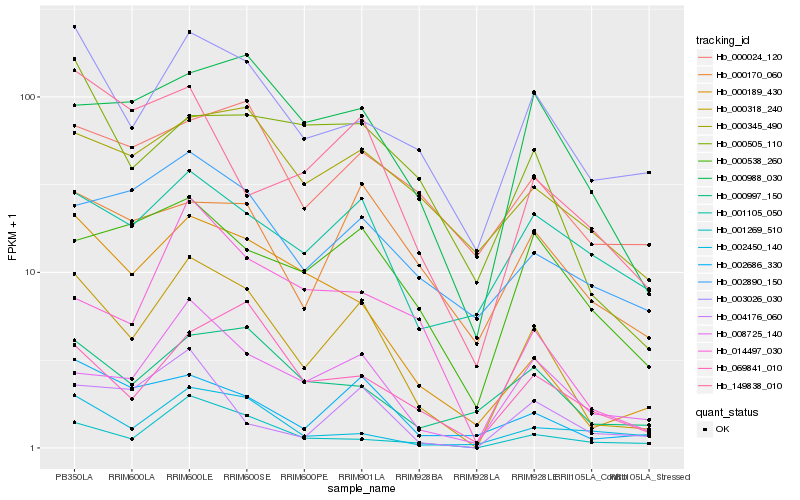
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002686_330 |
0.1848246582 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_008725_140 |
0.1858209597 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003026_030 |
0.1866558382 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 5 |
Hb_000189_430 |
0.1971916273 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_001269_510 |
0.2010516096 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 7 |
Hb_000997_150 |
0.2031463147 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000988_030 |
0.2060263702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_069841_010 |
0.2071536399 |
- |
- |
resistance protein [Quercus suber] |
| 10 |
Hb_000170_060 |
0.2165521363 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 11 |
Hb_000538_260 |
0.2171021881 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 12 |
Hb_002450_140 |
0.2214370252 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 13 |
Hb_004176_060 |
0.221553311 |
- |
- |
- |
| 14 |
Hb_000505_110 |
0.2219890056 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 15 |
Hb_002890_150 |
0.2229057102 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 16 |
Hb_149838_010 |
0.2229406954 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 17 |
Hb_000345_490 |
0.224135988 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 18 |
Hb_014497_030 |
0.2255991311 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 19 |
Hb_001105_050 |
0.2257121737 |
- |
- |
PREDICTED: uncharacterized protein LOC105642744 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000024_120 |
0.2259816338 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |