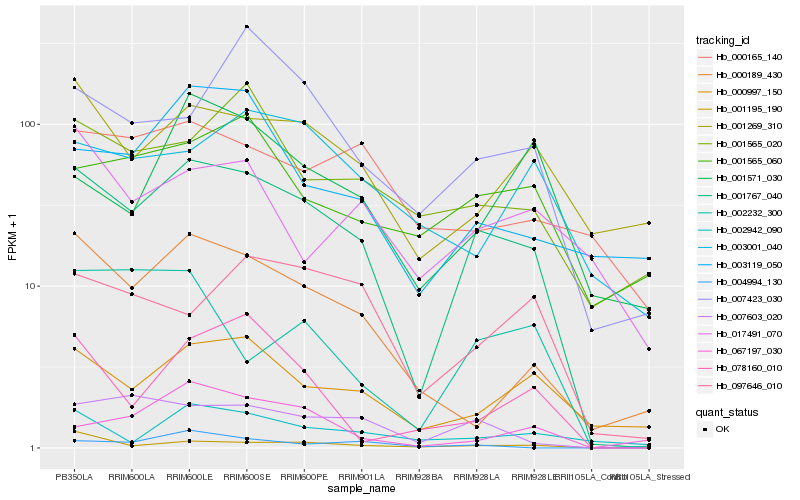
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001767_040 |
0.0 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 2 |
Hb_007603_020 |
0.1900814003 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 3 |
Hb_000997_150 |
0.2011181569 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000189_430 |
0.2089878792 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001269_310 |
0.2164208666 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 6 |
Hb_001195_190 |
0.2202869029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_097646_010 |
0.2226724711 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 8 |
Hb_001565_020 |
0.228194386 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial, partial [Jatropha curcas] |
| 9 |
Hb_001565_060 |
0.229868169 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_007423_030 |
0.2304968457 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |
| 11 |
Hb_003119_050 |
0.2308337287 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002942_090 |
0.2359001127 |
- |
- |
DNAJ heat shock N-terminal domain-containing protein, putative [Theobroma cacao] |
| 13 |
Hb_002232_300 |
0.2389500781 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 14 |
Hb_000165_140 |
0.2389757841 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 15 |
Hb_078160_010 |
0.241988998 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 16 |
Hb_001571_030 |
0.2437907653 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003001_040 |
0.2440988507 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 18 |
Hb_017491_070 |
0.2451235703 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_067197_030 |
0.2457389581 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004994_130 |
0.247849606 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |