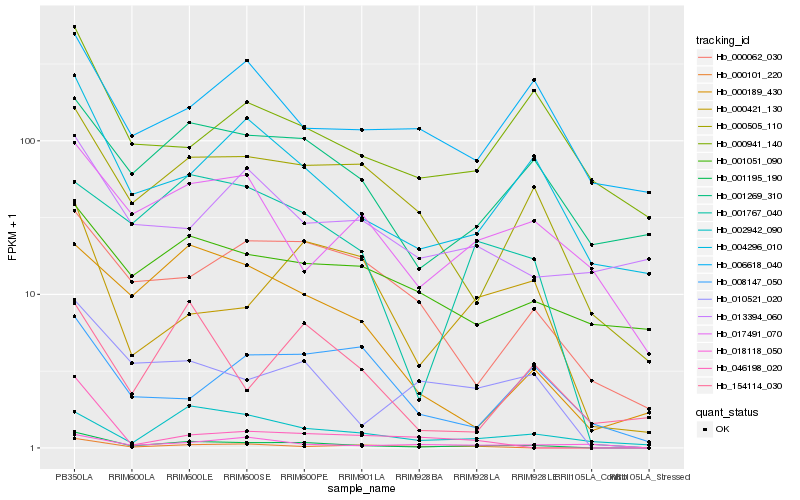
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010521_020 |
0.1920035587 |
- |
- |
- |
| 3 |
Hb_004296_010 |
0.2053277316 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 4 |
Hb_000505_110 |
0.2055884785 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 5 |
Hb_001767_040 |
0.2202869029 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 6 |
Hb_001269_310 |
0.2235399895 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 7 |
Hb_000421_130 |
0.2323556489 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000101_220 |
0.235507137 |
- |
- |
anthranilate synthase component I, putative [Ricinus communis] |
| 9 |
Hb_000062_030 |
0.2421916346 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000189_430 |
0.2461703596 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000941_140 |
0.2489211905 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 12 |
Hb_002942_090 |
0.2505940309 |
- |
- |
DNAJ heat shock N-terminal domain-containing protein, putative [Theobroma cacao] |
| 13 |
Hb_001051_090 |
0.2509613124 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 14 |
Hb_017491_070 |
0.2520000401 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_018118_050 |
0.2620992296 |
- |
- |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 16 |
Hb_154114_030 |
0.2670175514 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_006618_040 |
0.2684452622 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 18 |
Hb_008147_050 |
0.270785082 |
- |
- |
PREDICTED: uncharacterized protein LOC105646845 [Jatropha curcas] |
| 19 |
Hb_013394_060 |
0.2712042481 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 20 |
Hb_046198_020 |
0.2717351953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |