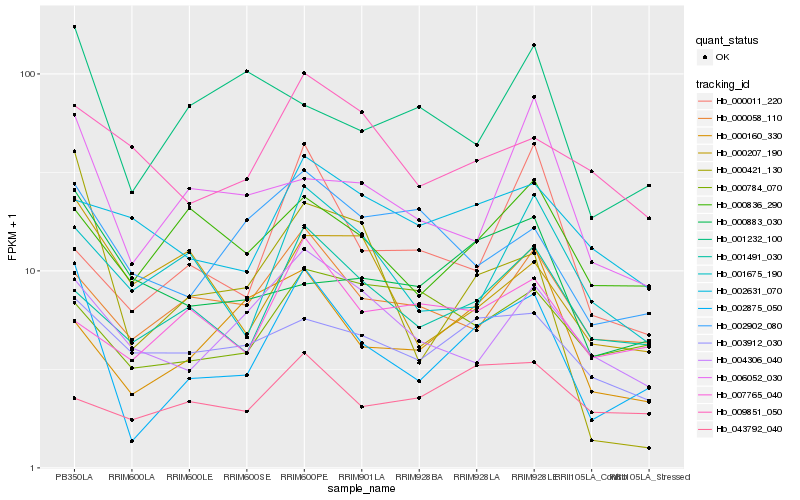
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002875_050 |
0.0 |
- |
- |
PREDICTED: proteoglycan 4 [Jatropha curcas] |
| 2 |
Hb_001491_030 |
0.2121726933 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000421_130 |
0.2150386757 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_000058_110 |
0.2167807065 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004306_040 |
0.2196574966 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 6 |
Hb_001675_190 |
0.2284733091 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 7 |
Hb_003912_030 |
0.2294931928 |
- |
- |
PREDICTED: uncharacterized protein LOC105638204 [Jatropha curcas] |
| 8 |
Hb_006052_030 |
0.2308730414 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A [Jatropha curcas] |
| 9 |
Hb_000160_330 |
0.2312104762 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_000836_290 |
0.2312170352 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_002902_080 |
0.2320259212 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 12 |
Hb_000883_030 |
0.2394188423 |
- |
- |
PREDICTED: uncharacterized protein LOC105636683 [Jatropha curcas] |
| 13 |
Hb_009851_050 |
0.2427705868 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 14 |
Hb_001232_100 |
0.2432171318 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 15 |
Hb_007765_040 |
0.24551494 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 16 |
Hb_000784_070 |
0.245944915 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000207_190 |
0.2460737548 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 18 |
Hb_000011_220 |
0.2471958293 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 19 |
Hb_002631_070 |
0.2494383447 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 20 |
Hb_043792_040 |
0.2499399364 |
- |
- |
hypothetical protein, partial [Rosa rugosa] |