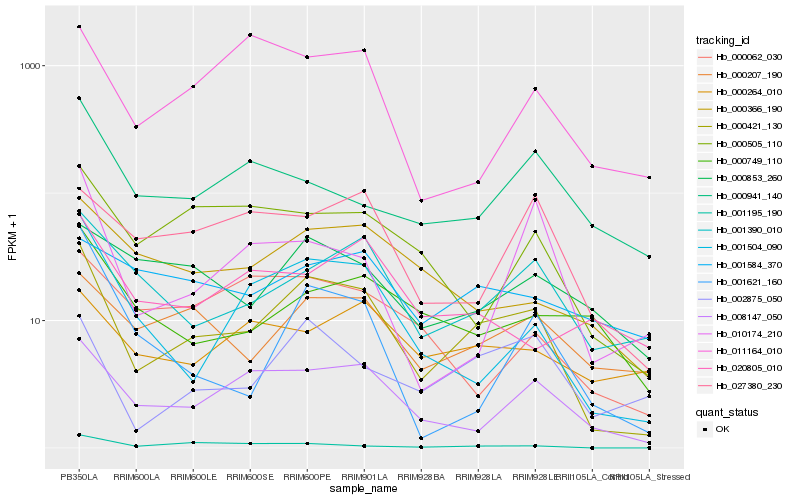
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000421_130 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_008147_050 |
0.1966544329 |
- |
- |
PREDICTED: uncharacterized protein LOC105646845 [Jatropha curcas] |
| 3 |
Hb_001504_090 |
0.2096155572 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_002875_050 |
0.2150386757 |
- |
- |
PREDICTED: proteoglycan 4 [Jatropha curcas] |
| 5 |
Hb_001390_010 |
0.2159379892 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 6 |
Hb_000207_190 |
0.2262841117 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 7 |
Hb_001195_190 |
0.2323556489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000366_190 |
0.2345494837 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 9 |
Hb_000505_110 |
0.2452561395 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 10 |
Hb_000749_110 |
0.2471963403 |
- |
- |
PREDICTED: choline transporter-like protein 2 [Jatropha curcas] |
| 11 |
Hb_000853_260 |
0.2501317619 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 12 |
Hb_027380_230 |
0.2514109102 |
- |
- |
PREDICTED: CSC1-like protein ERD4 [Jatropha curcas] |
| 13 |
Hb_001621_160 |
0.2517969446 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000264_010 |
0.2561558354 |
- |
- |
PREDICTED: uncharacterized protein LOC105649505 [Jatropha curcas] |
| 15 |
Hb_011164_010 |
0.2587468788 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 16 |
Hb_000941_140 |
0.2590472634 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 17 |
Hb_020805_010 |
0.2593125347 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_010174_210 |
0.2598763682 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000062_030 |
0.2614681755 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001584_370 |
0.2640316255 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |