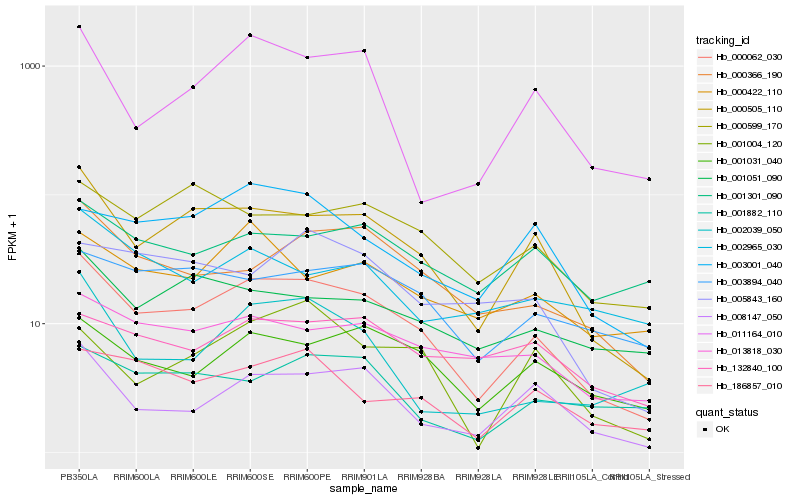
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_030 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000505_110 |
0.0992470104 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 3 |
Hb_000366_190 |
0.1485192877 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 4 |
Hb_008147_050 |
0.1492356501 |
- |
- |
PREDICTED: uncharacterized protein LOC105646845 [Jatropha curcas] |
| 5 |
Hb_001031_040 |
0.1579019421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_013818_030 |
0.1581046588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_186857_010 |
0.1632398689 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 8 |
Hb_011164_010 |
0.1677349459 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 9 |
Hb_003894_040 |
0.1699291393 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 10 |
Hb_001051_090 |
0.1741998324 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 11 |
Hb_000599_170 |
0.1748623405 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 12 |
Hb_000422_110 |
0.1768952272 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 13 |
Hb_003001_040 |
0.1769306503 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 14 |
Hb_002039_050 |
0.1819826861 |
- |
- |
PREDICTED: U-box domain-containing protein 40 [Jatropha curcas] |
| 15 |
Hb_001301_090 |
0.1830094846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005843_160 |
0.1838759798 |
- |
- |
PREDICTED: uncharacterized protein LOC105112024 [Populus euphratica] |
| 17 |
Hb_132840_100 |
0.1844644781 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 18 |
Hb_001882_110 |
0.1878969753 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002965_030 |
0.1884627433 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 20 |
Hb_001004_120 |
0.1885165622 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |