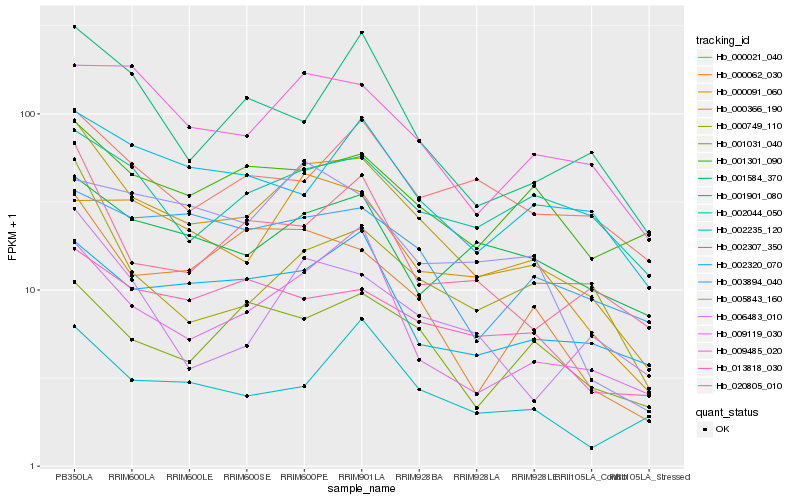
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000366_190 |
0.0 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 2 |
Hb_009119_030 |
0.144189061 |
- |
- |
GMFP4 [Medicago truncatula] |
| 3 |
Hb_020805_010 |
0.1478775185 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000062_030 |
0.1485192877 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 5 |
Hb_009485_020 |
0.1494690743 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 6 |
Hb_002320_070 |
0.1542607636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000749_110 |
0.1575877817 |
- |
- |
PREDICTED: choline transporter-like protein 2 [Jatropha curcas] |
| 8 |
Hb_006483_010 |
0.1585303534 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_013818_030 |
0.1587848284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001901_080 |
0.1610519783 |
- |
- |
PREDICTED: xylose isomerase [Jatropha curcas] |
| 11 |
Hb_003894_040 |
0.1618987249 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 12 |
Hb_001031_040 |
0.1626077306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001584_370 |
0.1656616138 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 14 |
Hb_000091_060 |
0.1662027842 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 15 |
Hb_002307_350 |
0.1669110397 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 16 |
Hb_002044_050 |
0.1679739654 |
- |
- |
- |
| 17 |
Hb_002235_120 |
0.1696268242 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 18 |
Hb_001301_090 |
0.17065914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000021_040 |
0.1709082419 |
- |
- |
PREDICTED: villin-3-like isoform X1 [Populus euphratica] |
| 20 |
Hb_005843_160 |
0.1709289334 |
- |
- |
PREDICTED: uncharacterized protein LOC105112024 [Populus euphratica] |