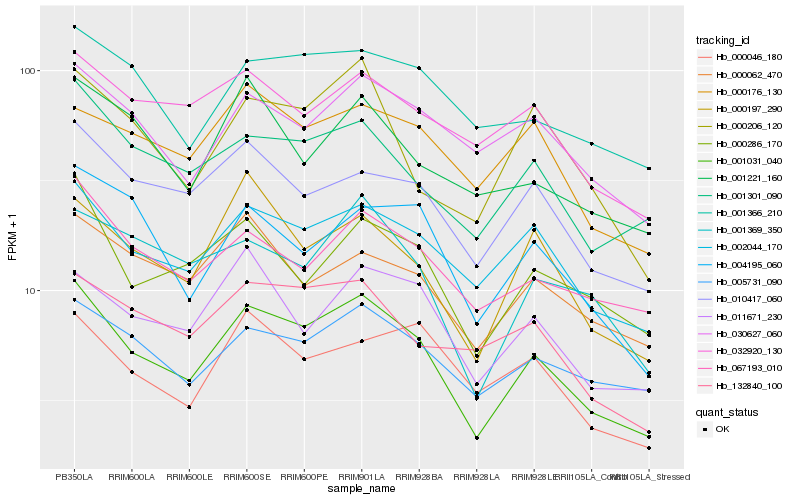
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001031_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002044_170 |
0.0955198973 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 3 |
Hb_000197_290 |
0.106010827 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 4 |
Hb_010417_060 |
0.106932965 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 5 |
Hb_001301_090 |
0.1070421351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000286_170 |
0.1110723658 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 7 |
Hb_011671_230 |
0.1121995889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005731_090 |
0.1131642528 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 9 |
Hb_001369_350 |
0.1134254532 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 10 |
Hb_067193_010 |
0.1152962546 |
- |
- |
- |
| 11 |
Hb_000206_120 |
0.1166707793 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_004195_060 |
0.1181728278 |
- |
- |
PREDICTED: uncharacterized protein LOC105803780 isoform X1 [Gossypium raimondii] |
| 13 |
Hb_030627_060 |
0.1183924878 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 14 |
Hb_132840_100 |
0.118706727 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 15 |
Hb_000062_470 |
0.1201226278 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 16 |
Hb_001366_210 |
0.1205861756 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 17 |
Hb_000176_130 |
0.1223270435 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 18 |
Hb_032920_130 |
0.1240876317 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 19 |
Hb_000046_180 |
0.1252492414 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |
| 20 |
Hb_001221_160 |
0.1256904119 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |