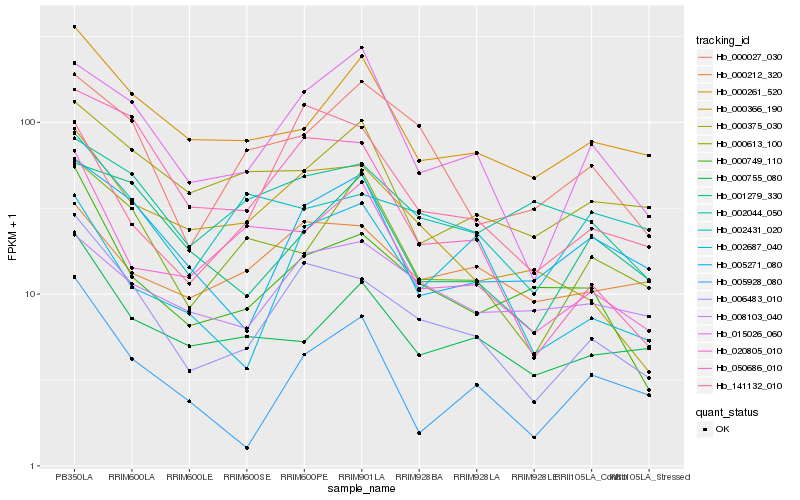
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006483_010 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000749_110 |
0.1546581035 |
- |
- |
PREDICTED: choline transporter-like protein 2 [Jatropha curcas] |
| 3 |
Hb_000366_190 |
0.1585303534 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 4 |
Hb_015026_060 |
0.1676768547 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 5 |
Hb_050686_010 |
0.1681293658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000261_520 |
0.1755048158 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 7 |
Hb_005271_080 |
0.1776551625 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 8 |
Hb_000027_030 |
0.1778647136 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_000755_080 |
0.1804930173 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 10 |
Hb_020805_010 |
0.1829757864 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_002431_020 |
0.1833600337 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 12 |
Hb_000613_100 |
0.1836331083 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 8 [Jatropha curcas] |
| 13 |
Hb_141132_010 |
0.1848513252 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |
| 14 |
Hb_000212_320 |
0.1865474165 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 15 |
Hb_001279_330 |
0.1867711302 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 16 |
Hb_008103_040 |
0.1921397925 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_005928_080 |
0.1930588837 |
- |
- |
PREDICTED: probable galacturonosyltransferase 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002687_040 |
0.1933598402 |
- |
- |
PREDICTED: alpha-galactosidase 1-like [Jatropha curcas] |
| 19 |
Hb_002044_050 |
0.1942137854 |
- |
- |
- |
| 20 |
Hb_000375_030 |
0.1954823872 |
- |
- |
- |