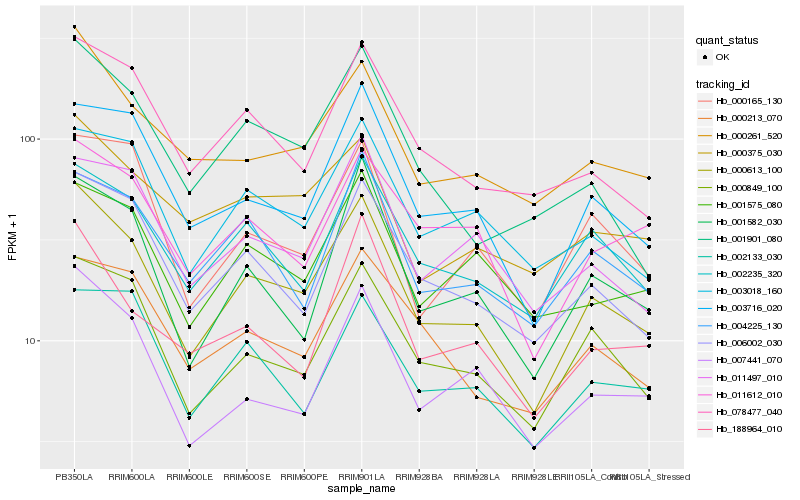
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000613_100 |
0.0 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 8 [Jatropha curcas] |
| 2 |
Hb_000849_100 |
0.0878764055 |
- |
- |
PREDICTED: uncharacterized protein LOC105644134 [Jatropha curcas] |
| 3 |
Hb_006002_030 |
0.0937707739 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003716_020 |
0.0978356577 |
- |
- |
PREDICTED: putative vesicle-associated membrane protein 726 [Jatropha curcas] |
| 5 |
Hb_001582_030 |
0.1013377595 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 6 |
Hb_002235_320 |
0.1021896664 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000261_520 |
0.1029010655 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 8 |
Hb_078477_040 |
0.1049129735 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 9 |
Hb_007441_070 |
0.1065673748 |
- |
- |
hypothetical protein PRUPE_ppa004494mg [Prunus persica] |
| 10 |
Hb_003018_160 |
0.1084476453 |
- |
- |
PREDICTED: uncharacterized protein LOC105628876 [Jatropha curcas] |
| 11 |
Hb_001901_080 |
0.1103110956 |
- |
- |
PREDICTED: xylose isomerase [Jatropha curcas] |
| 12 |
Hb_000165_130 |
0.1122310208 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 13 |
Hb_011497_010 |
0.1124925794 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 14 |
Hb_000213_070 |
0.1132737283 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004225_130 |
0.1135462575 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000375_030 |
0.1137465699 |
- |
- |
- |
| 17 |
Hb_188964_010 |
0.1145246906 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 18 |
Hb_011612_010 |
0.1162814595 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 19 |
Hb_001575_080 |
0.1174427076 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 20 |
Hb_002133_030 |
0.1178969349 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |