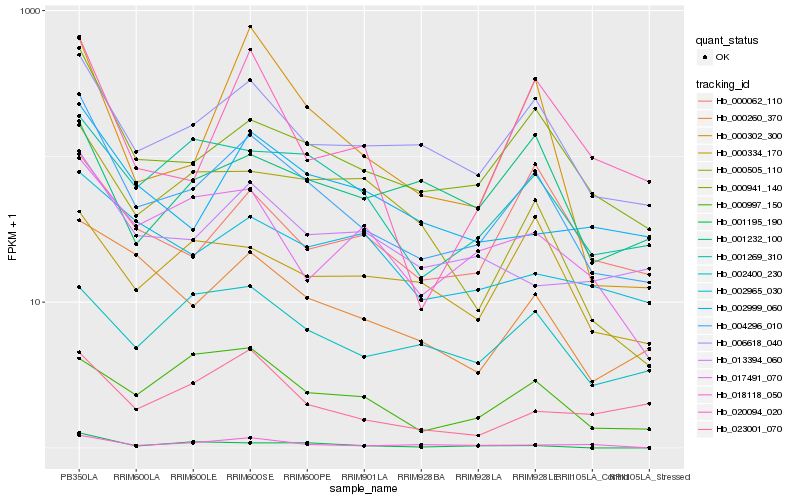
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004296_010 |
0.0 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 2 |
Hb_000941_140 |
0.1110565398 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 3 |
Hb_006618_040 |
0.1292061786 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 4 |
Hb_000260_370 |
0.164527457 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 5 |
Hb_001269_310 |
0.1754364379 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 6 |
Hb_002999_060 |
0.1780046329 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 7 |
Hb_013394_060 |
0.1809708517 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_017491_070 |
0.1911628368 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_020094_020 |
0.1924189338 |
- |
- |
PREDICTED: 17.1 kDa class II heat shock protein-like [Pyrus x bretschneideri] |
| 10 |
Hb_000302_300 |
0.1932978788 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 11 |
Hb_002965_030 |
0.1939644908 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 12 |
Hb_000062_110 |
0.1949612064 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 13 |
Hb_000505_110 |
0.1971405284 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 14 |
Hb_018118_050 |
0.1979856548 |
- |
- |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 15 |
Hb_023001_070 |
0.203697721 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A-like [Eucalyptus grandis] |
| 16 |
Hb_002400_230 |
0.2038335411 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 17 |
Hb_001195_190 |
0.2053277316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001232_100 |
0.2099057382 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 19 |
Hb_000334_170 |
0.2118645842 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000997_150 |
0.2120491423 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |