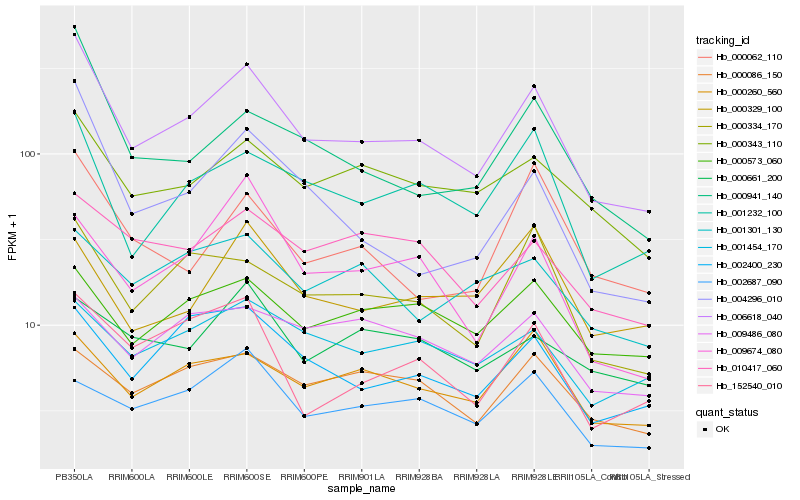
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006618_040 |
0.0 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 2 |
Hb_001232_100 |
0.1166931467 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 3 |
Hb_000334_170 |
0.1216245134 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000343_110 |
0.1240108778 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 5 |
Hb_000941_140 |
0.1279213707 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 6 |
Hb_002400_230 |
0.1285931761 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 7 |
Hb_004296_010 |
0.1292061786 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 8 |
Hb_152540_010 |
0.1336696401 |
- |
- |
- |
| 9 |
Hb_000260_560 |
0.1381760926 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 10 |
Hb_010417_060 |
0.1421657637 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 11 |
Hb_000062_110 |
0.1424736417 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 12 |
Hb_000573_060 |
0.1439608315 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 13 |
Hb_001454_170 |
0.1461725232 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 14 |
Hb_009674_080 |
0.1485150427 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 15 |
Hb_000086_150 |
0.1520414491 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001301_130 |
0.1544235632 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 17 |
Hb_000329_100 |
0.154837366 |
- |
- |
PREDICTED: uncharacterized protein LOC105643144 [Jatropha curcas] |
| 18 |
Hb_002687_090 |
0.1582674707 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 19 |
Hb_000661_200 |
0.1588084457 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009486_080 |
0.158970514 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |