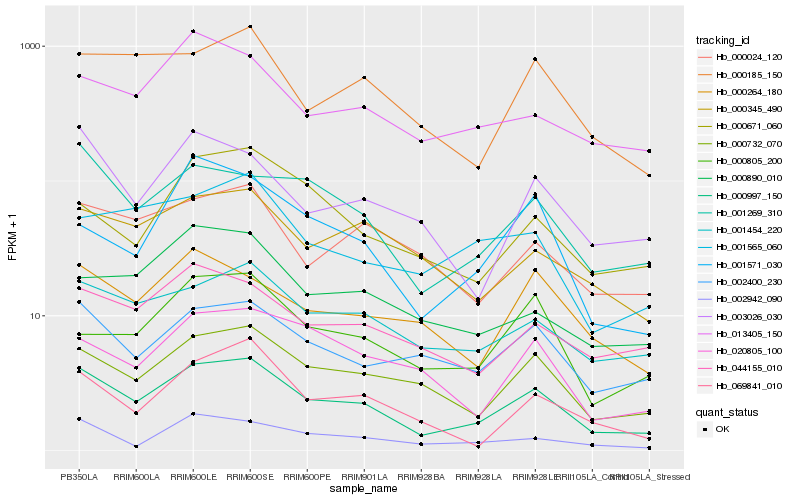
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_150 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000732_070 |
0.1176030655 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 3 |
Hb_001454_220 |
0.1228297228 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 4 |
Hb_001269_310 |
0.1289199966 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 5 |
Hb_002400_230 |
0.1313048808 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 6 |
Hb_044155_010 |
0.1417362499 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000185_150 |
0.1453987174 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000345_490 |
0.1483152592 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 9 |
Hb_003026_030 |
0.1491899434 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 10 |
Hb_013405_150 |
0.1508443001 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 11 |
Hb_002942_090 |
0.1513801552 |
- |
- |
DNAJ heat shock N-terminal domain-containing protein, putative [Theobroma cacao] |
| 12 |
Hb_000024_120 |
0.1516921696 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 13 |
Hb_069841_010 |
0.1523035515 |
- |
- |
resistance protein [Quercus suber] |
| 14 |
Hb_000805_200 |
0.1524410764 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000264_180 |
0.1525638881 |
- |
- |
PREDICTED: uncharacterized protein LOC100777318 [Glycine max] |
| 16 |
Hb_001571_030 |
0.1559457545 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001565_060 |
0.1564494092 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000671_060 |
0.1564863544 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_020805_100 |
0.1575056096 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 20 |
Hb_000890_010 |
0.1578686484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |