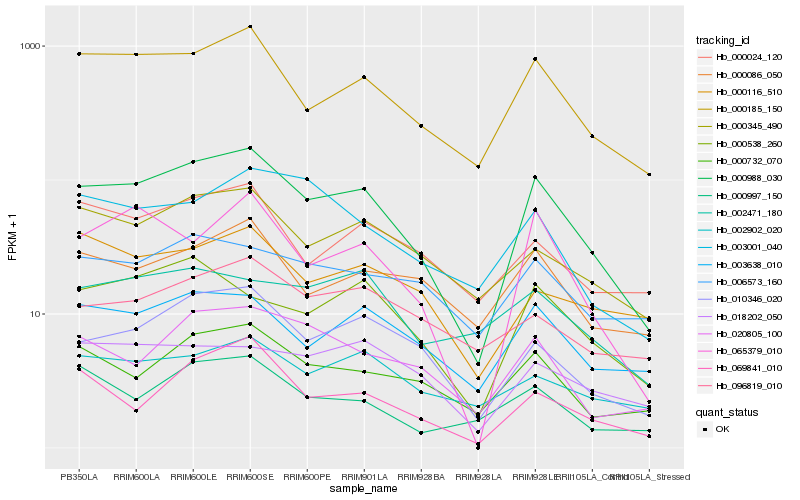
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000988_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000185_150 |
0.1092770702 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 3 |
Hb_010346_020 |
0.1273681703 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 4 |
Hb_065379_010 |
0.1409961437 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 5 |
Hb_003638_010 |
0.1418873065 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |
| 6 |
Hb_000732_070 |
0.144271561 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 7 |
Hb_000345_490 |
0.1457212775 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 8 |
Hb_000538_260 |
0.1464579549 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 9 |
Hb_020805_100 |
0.153898119 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 10 |
Hb_069841_010 |
0.1545500278 |
- |
- |
resistance protein [Quercus suber] |
| 11 |
Hb_000086_050 |
0.1562693208 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 12 |
Hb_000024_120 |
0.1596132144 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 13 |
Hb_003001_040 |
0.1601301135 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 14 |
Hb_002902_020 |
0.1621906993 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 15 |
Hb_018202_050 |
0.16374739 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A [Jatropha curcas] |
| 16 |
Hb_000997_150 |
0.1649693374 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_006573_160 |
0.1674201472 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 18 |
Hb_000116_510 |
0.1689583688 |
- |
- |
PREDICTED: RNA-binding protein 24-A-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_096819_010 |
0.1697868142 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 20 |
Hb_002471_180 |
0.1712028012 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |