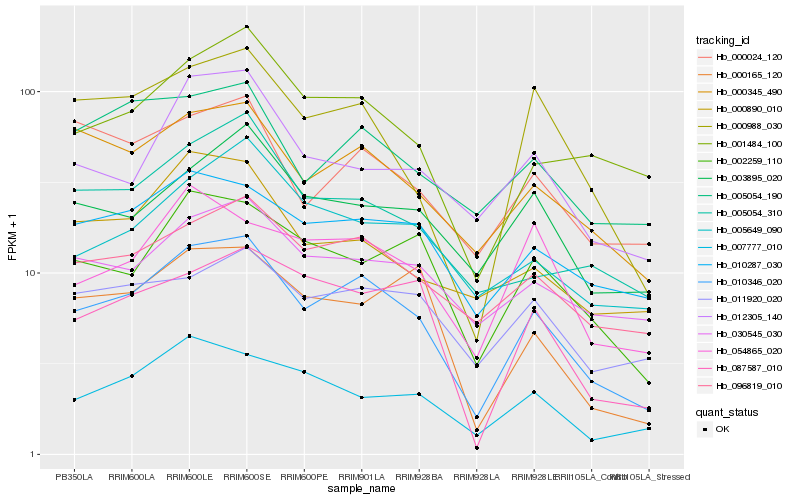
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010346_020 |
0.0 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 2 |
Hb_096819_010 |
0.1191448475 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 3 |
Hb_000345_490 |
0.1196074874 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 4 |
Hb_000988_030 |
0.1273681703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002259_110 |
0.1301557696 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 6 |
Hb_001484_100 |
0.131888207 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 7 |
Hb_010287_030 |
0.1326224699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007777_010 |
0.1370090293 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Tarenaya hassleriana] |
| 9 |
Hb_005054_310 |
0.1378249168 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 10 |
Hb_003895_020 |
0.1381052244 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 11 |
Hb_087587_010 |
0.1381915701 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 12 |
Hb_000165_120 |
0.1395212104 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 13 |
Hb_005649_090 |
0.1404413092 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 14 |
Hb_005054_190 |
0.1408636698 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 15 |
Hb_030545_030 |
0.1411484018 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 16 |
Hb_012305_140 |
0.1412150259 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000890_010 |
0.1420378029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000024_120 |
0.1427475549 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 19 |
Hb_054865_020 |
0.1428600883 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 20 |
Hb_011920_020 |
0.1430303622 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |