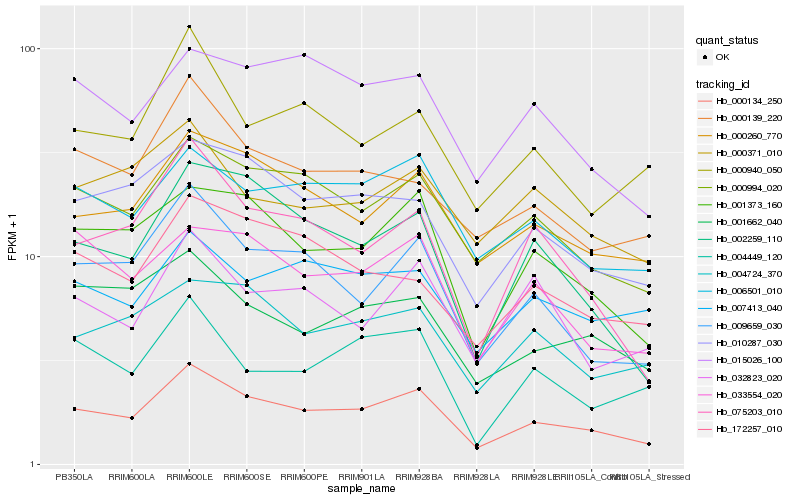
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000134_250 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_000994_020 |
0.0862352115 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_001373_160 |
0.0863540011 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 4 |
Hb_000260_770 |
0.0906258683 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 5 |
Hb_009659_030 |
0.0956209663 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 6 |
Hb_006501_010 |
0.097785283 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A isoform X1 [Populus euphratica] |
| 7 |
Hb_010287_030 |
0.0997119937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002259_110 |
0.1047229313 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 9 |
Hb_075203_010 |
0.1069291968 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_033554_020 |
0.1105408255 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 11 |
Hb_004724_370 |
0.1114847144 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 12 |
Hb_001662_040 |
0.1138646444 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 13 |
Hb_000139_220 |
0.1157879358 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_015026_100 |
0.1170665271 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 15 |
Hb_000940_050 |
0.1177649069 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 16 |
Hb_172257_010 |
0.1191946397 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 17 |
Hb_007413_040 |
0.1203020805 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 18 |
Hb_000371_010 |
0.1213606895 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 19 |
Hb_032823_020 |
0.1217353599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004449_120 |
0.1222028929 |
- |
- |
PREDICTED: uncharacterized protein LOC105631291 [Jatropha curcas] |