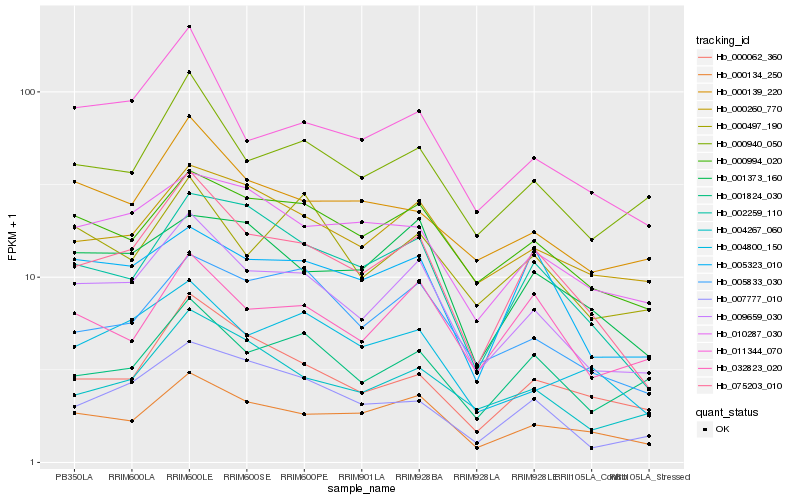
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009659_030 |
0.0 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 2 |
Hb_075203_010 |
0.0857627727 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000134_250 |
0.0956209663 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000994_020 |
0.0998223063 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_011344_070 |
0.1008712633 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 6 |
Hb_000940_050 |
0.1014035009 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 7 |
Hb_005833_030 |
0.1106061707 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 8 |
Hb_004267_060 |
0.1124657767 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_000260_770 |
0.1125134331 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 10 |
Hb_000139_220 |
0.1173068854 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002259_110 |
0.1192160593 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 12 |
Hb_032823_020 |
0.1202146178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000497_190 |
0.1207621988 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 14 |
Hb_001824_030 |
0.1215473847 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 15 |
Hb_004800_150 |
0.1217627074 |
transcription factor |
TF Family: SNF2 |
PREDICTED: ATP-dependent DNA helicase DDM1 [Jatropha curcas] |
| 16 |
Hb_010287_030 |
0.1219025724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007777_010 |
0.1226946217 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Tarenaya hassleriana] |
| 18 |
Hb_001373_160 |
0.1231344433 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 19 |
Hb_000062_360 |
0.1246815772 |
- |
- |
PREDICTED: DNA polymerase alpha subunit B [Jatropha curcas] |
| 20 |
Hb_005323_010 |
0.1265946905 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |