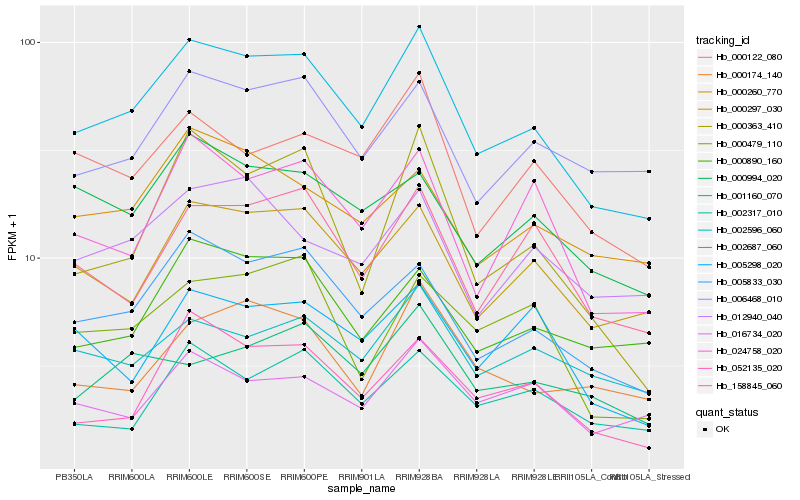
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002687_060 |
0.0 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 2 |
Hb_005833_030 |
0.0772791788 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 3 |
Hb_000297_030 |
0.0911356545 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_024758_020 |
0.1082739397 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 5 |
Hb_006468_010 |
0.1101152697 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_158845_060 |
0.1102386332 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000122_080 |
0.1109349128 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 8 |
Hb_000890_160 |
0.113268169 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 9 |
Hb_002596_060 |
0.113657076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000994_020 |
0.1143094549 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_052135_020 |
0.1151960932 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 12 |
Hb_000260_770 |
0.1159037153 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 13 |
Hb_012940_040 |
0.1186943759 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 14 |
Hb_016734_020 |
0.1205102839 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 15 |
Hb_002317_010 |
0.1208134473 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_000363_410 |
0.1231784379 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 17 |
Hb_000174_140 |
0.1236530901 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 18 |
Hb_000479_110 |
0.1243296435 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005298_020 |
0.1257812453 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 2 [Jatropha curcas] |
| 20 |
Hb_001160_070 |
0.1258905026 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |