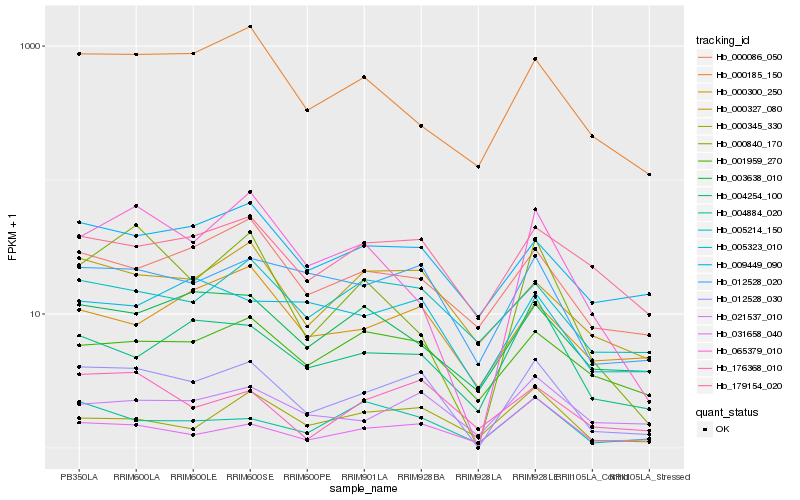
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012528_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_012528_020 |
0.1607965738 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Jatropha curcas] |
| 3 |
Hb_000327_080 |
0.1772714879 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 4 |
Hb_021537_010 |
0.1802982585 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 5 |
Hb_176368_010 |
0.1829772299 |
- |
- |
hypothetical protein POPTR_0019s00300g [Populus trichocarpa] |
| 6 |
Hb_004254_100 |
0.1858980562 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009449_090 |
0.1860398743 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 8 |
Hb_179154_020 |
0.1900928313 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 9 |
Hb_000086_050 |
0.1913967022 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 10 |
Hb_065379_010 |
0.1932661986 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 11 |
Hb_001959_270 |
0.1934457683 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_004884_020 |
0.1939709117 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X2 [Jatropha curcas] |
| 13 |
Hb_005214_150 |
0.195243766 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 14 |
Hb_000185_150 |
0.1974133426 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000840_170 |
0.1980256101 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 16 |
Hb_000345_330 |
0.1988018911 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance-like protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_031658_040 |
0.1994983242 |
- |
- |
- |
| 18 |
Hb_000300_250 |
0.2011856142 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 19 |
Hb_005323_010 |
0.2014684875 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |
| 20 |
Hb_003638_010 |
0.2014734262 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |