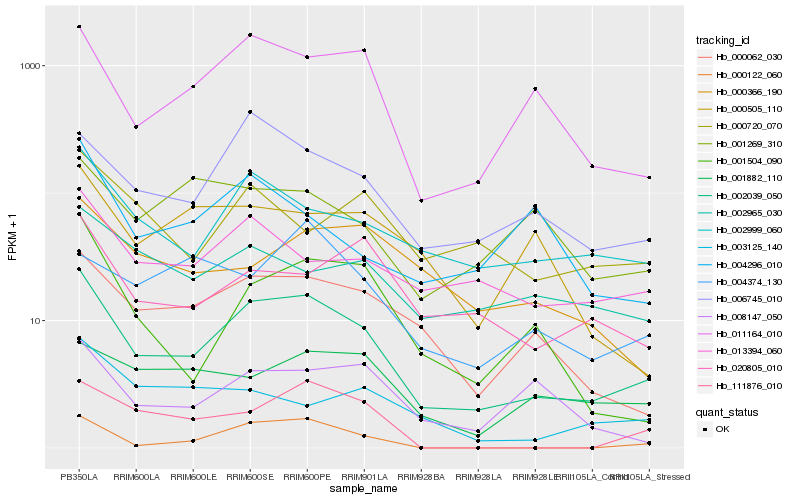
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002039_050 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 40 [Jatropha curcas] |
| 2 |
Hb_011164_010 |
0.1778286743 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 3 |
Hb_002999_060 |
0.1792186227 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 4 |
Hb_006745_010 |
0.180368169 |
- |
- |
PREDICTED: uncharacterized protein LOC105640095 [Jatropha curcas] |
| 5 |
Hb_000062_030 |
0.1819826861 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001504_090 |
0.2033801019 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_013394_060 |
0.2053433089 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_020805_010 |
0.2148046994 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_008147_050 |
0.2164154309 |
- |
- |
PREDICTED: uncharacterized protein LOC105646845 [Jatropha curcas] |
| 10 |
Hb_000122_060 |
0.2180771894 |
- |
- |
PREDICTED: protein LURP-one-related 14 [Jatropha curcas] |
| 11 |
Hb_002965_030 |
0.2194352908 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 12 |
Hb_001882_110 |
0.2237702494 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000720_070 |
0.2286069769 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 14 |
Hb_111876_010 |
0.2338913568 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 15 |
Hb_000366_190 |
0.2344593839 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 16 |
Hb_004374_130 |
0.2360485139 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 17 |
Hb_004296_010 |
0.236055269 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 18 |
Hb_003125_140 |
0.2361752846 |
- |
- |
PREDICTED: uncharacterized protein LOC105648796 [Jatropha curcas] |
| 19 |
Hb_000505_110 |
0.2370192461 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 20 |
Hb_001269_310 |
0.239182073 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |