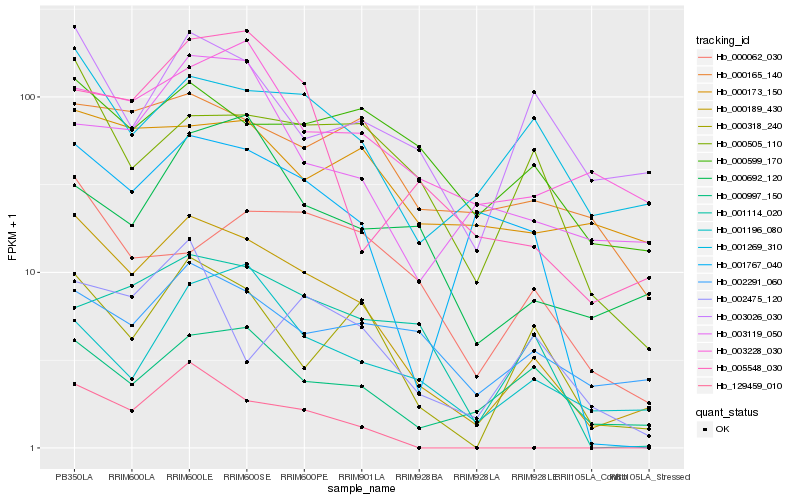
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_430 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000318_240 |
0.1971916273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_129459_010 |
0.1982096849 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 4 |
Hb_001767_040 |
0.2089878792 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 5 |
Hb_001269_310 |
0.2093519578 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 6 |
Hb_005548_030 |
0.213408145 |
- |
- |
hypothetical protein JCGZ_05606 [Jatropha curcas] |
| 7 |
Hb_000997_150 |
0.21417211 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000505_110 |
0.2154790821 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 9 |
Hb_000165_140 |
0.220826785 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 10 |
Hb_003119_050 |
0.2210177696 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001196_080 |
0.2213338367 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 12 |
Hb_003026_030 |
0.2225658142 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 13 |
Hb_002291_060 |
0.2255107785 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_000062_030 |
0.2281802032 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000692_120 |
0.2292956681 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 16 |
Hb_001114_020 |
0.229368157 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 17 |
Hb_003228_030 |
0.2301666433 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_000173_150 |
0.2307034814 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 19 |
Hb_002475_120 |
0.2309845889 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 20 |
Hb_000599_170 |
0.2309877731 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |