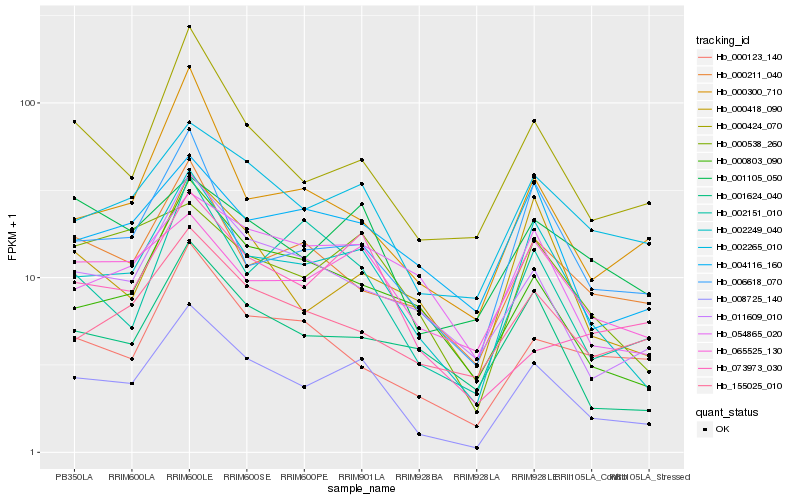
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_140 |
0.0 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002249_040 |
0.0991470451 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1-like [Jatropha curcas] |
| 3 |
Hb_002265_010 |
0.1349345543 |
- |
- |
ef-hand calcium binding protein, putative [Ricinus communis] |
| 4 |
Hb_000424_070 |
0.1514191698 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 5 |
Hb_000538_260 |
0.1590339674 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 6 |
Hb_011609_010 |
0.1592964121 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 7 |
Hb_006618_070 |
0.16215269 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000803_090 |
0.1650966566 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 9 |
Hb_001624_040 |
0.1681789097 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_065525_130 |
0.1700528211 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_155025_010 |
0.1717462463 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 12 |
Hb_001105_050 |
0.1724181785 |
- |
- |
PREDICTED: uncharacterized protein LOC105642744 isoform X1 [Jatropha curcas] |
| 13 |
Hb_073973_030 |
0.1734008163 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000418_090 |
0.1745511059 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_002151_010 |
0.1757528329 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 16 |
Hb_054865_020 |
0.1764467454 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 17 |
Hb_004116_160 |
0.1771669693 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000123_140 |
0.1782592704 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 19 |
Hb_000211_040 |
0.179002179 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 20 |
Hb_000300_710 |
0.1793943568 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |