| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
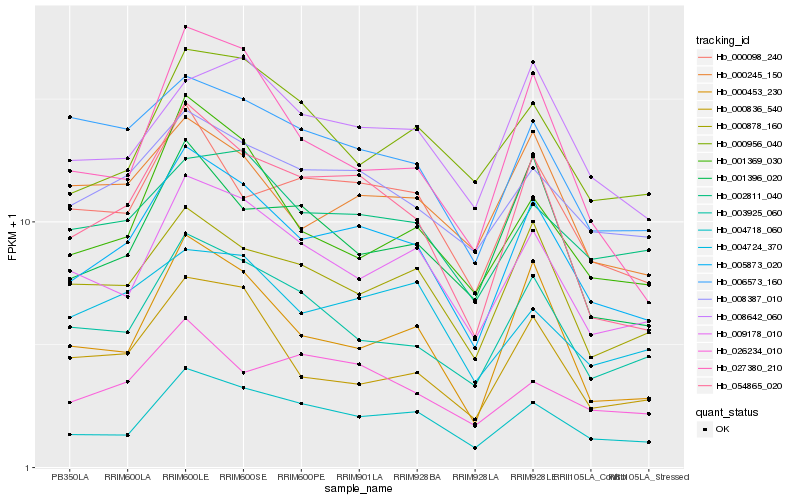
Hb_005873_020 |
0.0 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 2 |
Hb_054865_020 |
0.0733635412 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 3 |
Hb_004718_060 |
0.0771706128 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_009178_010 |
0.0889847054 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 5 |
Hb_008387_010 |
0.0929818101 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 6 |
Hb_003925_060 |
0.0960982696 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000878_160 |
0.0974971634 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 8 |
Hb_000098_240 |
0.0981819588 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 9 |
Hb_000453_230 |
0.1034086222 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 10 |
Hb_001369_030 |
0.1076264282 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 11 |
Hb_002811_040 |
0.1078133247 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 12 |
Hb_000836_540 |
0.110511861 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 13 |
Hb_000245_150 |
0.1108374245 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 14 |
Hb_004724_370 |
0.1108507159 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 15 |
Hb_001396_020 |
0.11161135 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 16 |
Hb_008642_060 |
0.1128332119 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 17 |
Hb_026234_010 |
0.1131736231 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 18 |
Hb_000956_040 |
0.1137135789 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 19 |
Hb_027380_210 |
0.1138018553 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 20 |
Hb_006573_160 |
0.1153902786 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |