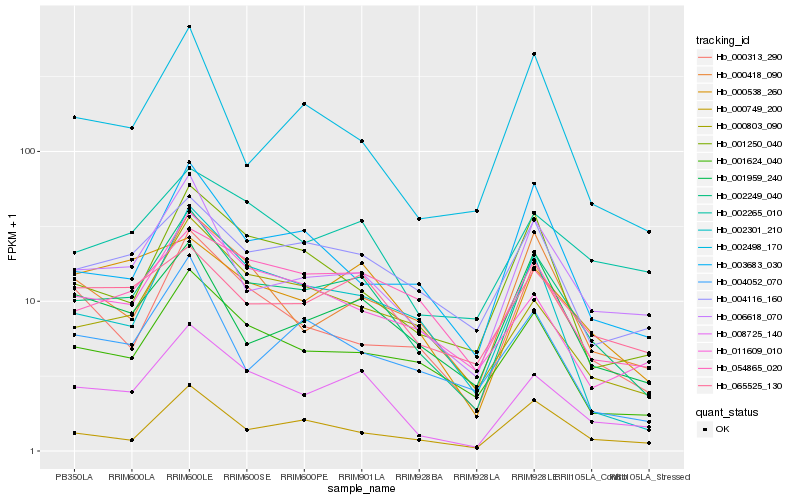
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002249_040 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1-like [Jatropha curcas] |
| 2 |
Hb_008725_140 |
0.0991470451 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_006618_070 |
0.1201392414 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001624_040 |
0.1259260639 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004116_160 |
0.1354131231 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002301_210 |
0.138366242 |
- |
- |
PREDICTED: receptor-like protein kinase At5g59670 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000749_200 |
0.1423243135 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000803_090 |
0.1426841007 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 9 |
Hb_001250_040 |
0.1440833139 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 10 |
Hb_000418_090 |
0.1443586513 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_003683_030 |
0.1447459369 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 12 |
Hb_001959_240 |
0.1454271172 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit B, chloroplastic/mitochondrial [Jatropha curcas] |
| 13 |
Hb_054865_020 |
0.1465803067 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 14 |
Hb_000313_290 |
0.1511402463 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_002265_010 |
0.1542268399 |
- |
- |
ef-hand calcium binding protein, putative [Ricinus communis] |
| 16 |
Hb_002498_170 |
0.1553081712 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_065525_130 |
0.1563376962 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_004052_070 |
0.1564200304 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 19 |
Hb_000538_260 |
0.1573225476 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 20 |
Hb_011609_010 |
0.1578472422 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |