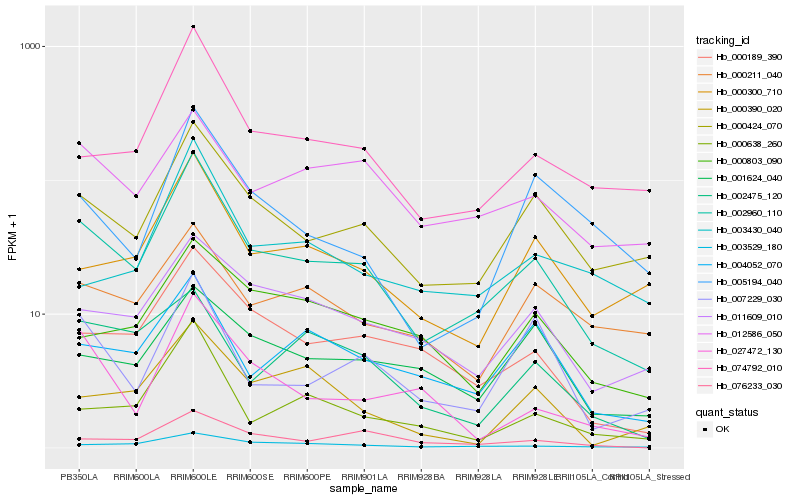
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002960_110 |
0.0 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000424_070 |
0.1368401134 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 3 |
Hb_074792_010 |
0.1530317641 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 4 |
Hb_000189_390 |
0.156469273 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003529_180 |
0.1651072857 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 6 |
Hb_000300_710 |
0.1720788258 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 7 |
Hb_000638_260 |
0.1735426005 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 8 |
Hb_011609_010 |
0.1767417094 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 9 |
Hb_004052_070 |
0.1846739241 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 10 |
Hb_076233_030 |
0.1879666634 |
- |
- |
PREDICTED: phosphoserine phosphatase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_007229_030 |
0.1880675715 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_012586_050 |
0.1892738621 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Prunus mume] |
| 13 |
Hb_005194_040 |
0.1926530415 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_027472_130 |
0.1969638417 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 15 |
Hb_003430_040 |
0.1975112968 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 16 |
Hb_000211_040 |
0.1983917185 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 17 |
Hb_002475_120 |
0.2008793403 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 18 |
Hb_001624_040 |
0.2028703363 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000390_020 |
0.2041656414 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 20 |
Hb_000803_090 |
0.2042623681 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |