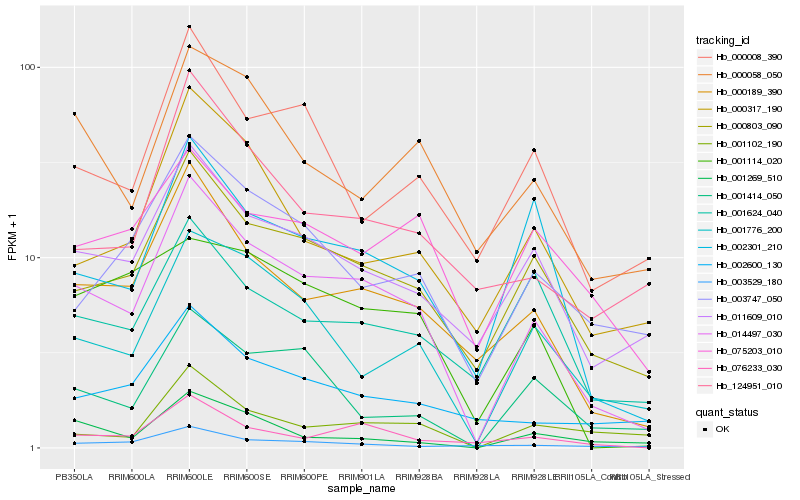
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014497_030 |
0.0 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 2 |
Hb_000189_390 |
0.1280873864 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000803_090 |
0.1323839716 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 4 |
Hb_011609_010 |
0.1527294178 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 5 |
Hb_001776_200 |
0.1599029183 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 6 |
Hb_002301_210 |
0.1670071342 |
- |
- |
PREDICTED: receptor-like protein kinase At5g59670 isoform X2 [Jatropha curcas] |
| 7 |
Hb_076233_030 |
0.1689404482 |
- |
- |
PREDICTED: phosphoserine phosphatase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003747_050 |
0.1770442691 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 9 |
Hb_000317_190 |
0.1784017909 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 10 |
Hb_124951_010 |
0.179610534 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 11 |
Hb_000008_390 |
0.1809127554 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 12 |
Hb_000058_050 |
0.1827263304 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 13 |
Hb_002600_130 |
0.1836891958 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 14 |
Hb_001414_050 |
0.1854850032 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 15 |
Hb_001114_020 |
0.1924806895 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 16 |
Hb_001269_510 |
0.1929467031 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 17 |
Hb_003529_180 |
0.1941861768 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 18 |
Hb_001102_190 |
0.1952934481 |
- |
- |
PREDICTED: uncharacterized protein LOC105112724 [Populus euphratica] |
| 19 |
Hb_001624_040 |
0.1956030151 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_075203_010 |
0.1957847954 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |