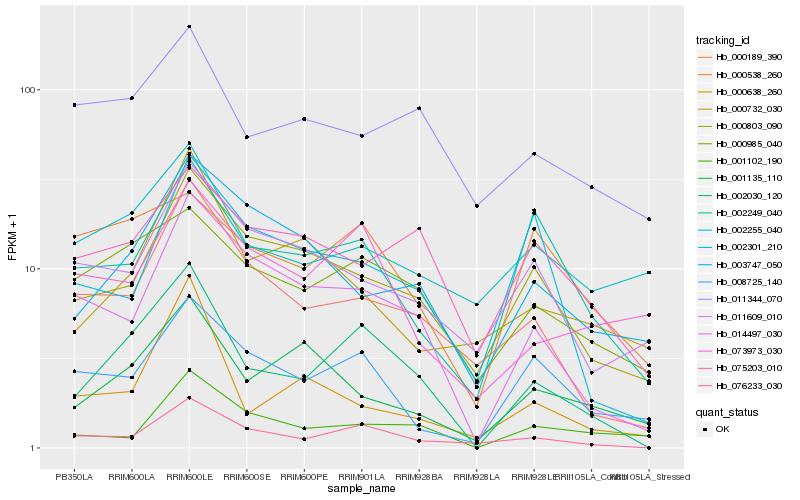
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002030_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_076233_030 |
0.1813402487 |
- |
- |
PREDICTED: phosphoserine phosphatase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_014497_030 |
0.2115913086 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 4 |
Hb_000189_390 |
0.2118509537 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000985_040 |
0.2143297892 |
transcription factor |
TF Family: Orphans |
B-box type zinc finger protein with CCT domain isoform 1 [Theobroma cacao] |
| 6 |
Hb_000803_090 |
0.2274989341 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 7 |
Hb_002249_040 |
0.232320589 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1-like [Jatropha curcas] |
| 8 |
Hb_008725_140 |
0.2407539948 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_073973_030 |
0.2470342573 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001135_110 |
0.2485306171 |
- |
- |
- |
| 11 |
Hb_000538_260 |
0.252327435 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 12 |
Hb_075203_010 |
0.2528283193 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_011344_070 |
0.2533337457 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 14 |
Hb_000638_260 |
0.2543176503 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 15 |
Hb_003747_050 |
0.2571759282 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 16 |
Hb_001102_190 |
0.2591966344 |
- |
- |
PREDICTED: uncharacterized protein LOC105112724 [Populus euphratica] |
| 17 |
Hb_000732_030 |
0.2601416058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002301_210 |
0.2601818422 |
- |
- |
PREDICTED: receptor-like protein kinase At5g59670 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002255_040 |
0.2613710648 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 20 |
Hb_011609_010 |
0.2624882707 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |