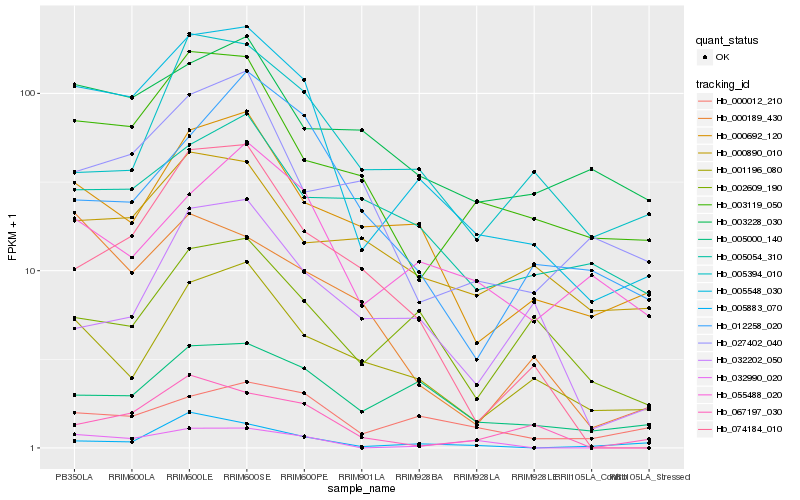
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005548_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_05606 [Jatropha curcas] |
| 2 |
Hb_000692_120 |
0.1696787766 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 3 |
Hb_003119_050 |
0.184387967 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001196_080 |
0.1860217043 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 5 |
Hb_005000_140 |
0.1928195391 |
- |
- |
PREDICTED: uncharacterized protein LOC105637876 isoform X1 [Jatropha curcas] |
| 6 |
Hb_074184_010 |
0.199126998 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 7 |
Hb_055488_020 |
0.1992481011 |
- |
- |
PREDICTED: uncharacterized protein LOC105641270 [Jatropha curcas] |
| 8 |
Hb_000012_210 |
0.2072534543 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002609_190 |
0.2077319817 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 10 |
Hb_005394_010 |
0.2101014104 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 11 |
Hb_067197_030 |
0.2107494422 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005054_310 |
0.2109163363 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 13 |
Hb_032990_020 |
0.2123370139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_027402_040 |
0.2125985794 |
- |
- |
Thioredoxin II, putative [Ricinus communis] |
| 15 |
Hb_003228_030 |
0.212815461 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_000189_430 |
0.213408145 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000890_010 |
0.2160964468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_012258_020 |
0.2166919598 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 19 |
Hb_032202_050 |
0.2172464988 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005883_070 |
0.2177965526 |
- |
- |
PREDICTED: probable myosin-binding protein 5 [Jatropha curcas] |