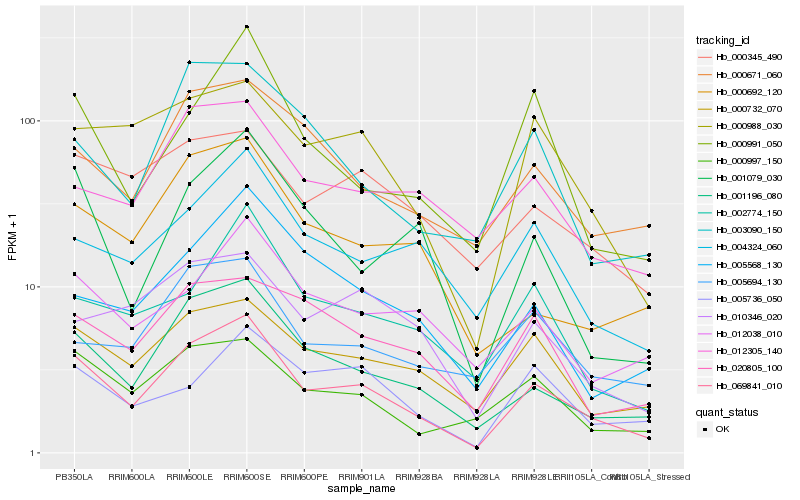
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_069841_010 |
0.0 |
- |
- |
resistance protein [Quercus suber] |
| 2 |
Hb_001196_080 |
0.1152671439 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 3 |
Hb_000732_070 |
0.1477398302 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 4 |
Hb_000997_150 |
0.1523035515 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000988_030 |
0.1545500278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000671_060 |
0.1575165472 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_001079_030 |
0.164974479 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Jatropha curcas] |
| 8 |
Hb_004324_060 |
0.167288993 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 9 |
Hb_000991_050 |
0.1681657903 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 10 |
Hb_020805_100 |
0.1689125885 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 11 |
Hb_000692_120 |
0.1690236345 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 12 |
Hb_012038_010 |
0.170383962 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 13 |
Hb_000345_490 |
0.1704445883 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 14 |
Hb_003090_150 |
0.1708743379 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 15 |
Hb_002774_150 |
0.1720444533 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 16 |
Hb_005736_050 |
0.1732207173 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 17 |
Hb_010346_020 |
0.1736111095 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 18 |
Hb_012305_140 |
0.1737771792 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005694_130 |
0.1761009202 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 20 |
Hb_005568_130 |
0.1762145336 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |