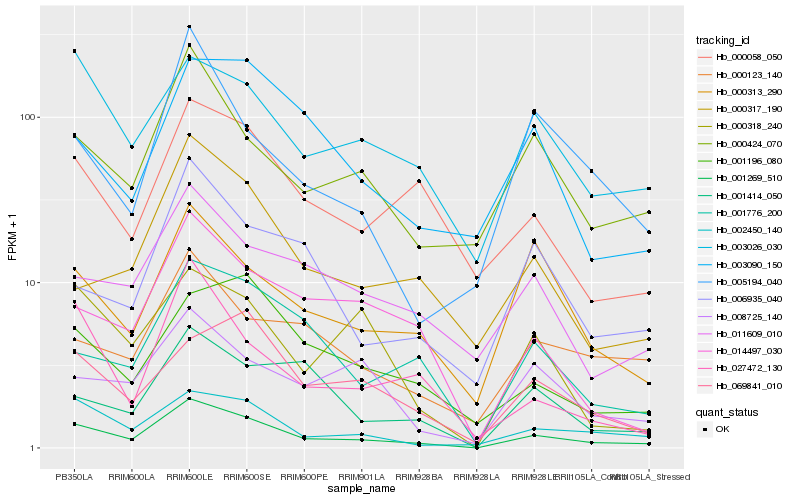
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_510 |
0.0 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 2 |
Hb_002450_140 |
0.1567742389 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 3 |
Hb_000424_070 |
0.1728298853 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 4 |
Hb_001776_200 |
0.1794960263 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 5 |
Hb_000058_050 |
0.1826281397 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 6 |
Hb_000313_290 |
0.1854458217 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_003026_030 |
0.1907329997 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 8 |
Hb_027472_130 |
0.1908865802 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 9 |
Hb_000123_140 |
0.1911970465 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 10 |
Hb_011609_010 |
0.1914605236 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 11 |
Hb_000317_190 |
0.1922045662 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 12 |
Hb_014497_030 |
0.1929467031 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 13 |
Hb_006935_040 |
0.1939329941 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 14 |
Hb_008725_140 |
0.1948044031 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001196_080 |
0.1972457282 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 16 |
Hb_001414_050 |
0.1979561781 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 17 |
Hb_069841_010 |
0.1988288947 |
- |
- |
resistance protein [Quercus suber] |
| 18 |
Hb_005194_040 |
0.2002617013 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000318_240 |
0.2010516096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003090_150 |
0.201308026 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |