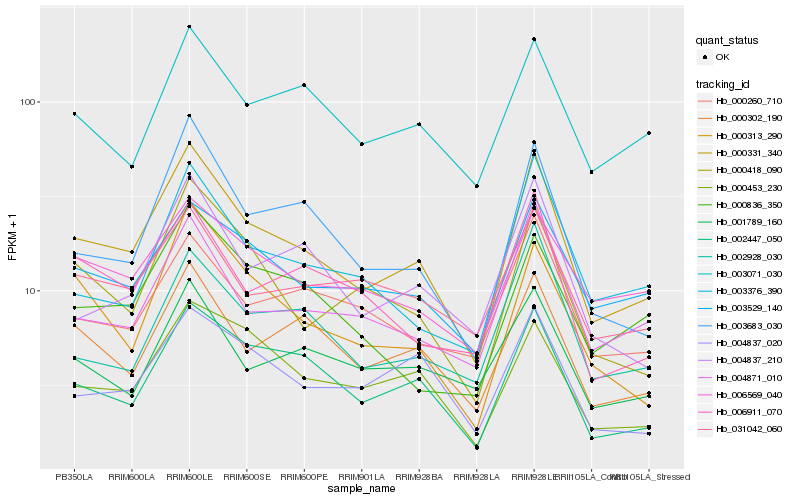
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640234 [Jatropha curcas] |
| 2 |
Hb_002447_050 |
0.0959947515 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_003683_030 |
0.1035994258 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 4 |
Hb_000302_190 |
0.1099309985 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |
| 5 |
Hb_003529_140 |
0.1140872797 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004871_010 |
0.1141170664 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 7 |
Hb_000418_090 |
0.115089999 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000453_230 |
0.1176270137 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_004837_020 |
0.1217893647 |
- |
- |
PREDICTED: uncharacterized protein LOC105648271 [Jatropha curcas] |
| 10 |
Hb_004837_210 |
0.1225133177 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 11 |
Hb_000313_290 |
0.1243305275 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_006911_070 |
0.1244774297 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 13 |
Hb_003376_390 |
0.1246757481 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002928_030 |
0.1258024962 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 15 |
Hb_003071_030 |
0.1261456526 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 16 |
Hb_031042_060 |
0.12616236 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 17 |
Hb_006569_040 |
0.1275087931 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 18 |
Hb_000260_710 |
0.127674972 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001789_160 |
0.129187082 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 20 |
Hb_000836_350 |
0.1320818148 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |