| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
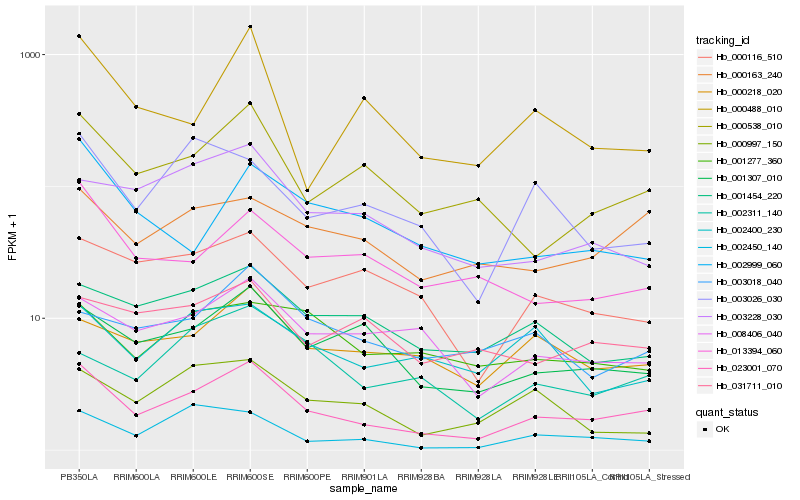
Hb_023001_070 |
0.0 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A-like [Eucalyptus grandis] |
| 2 |
Hb_000538_010 |
0.1646106652 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001307_010 |
0.1650536474 |
- |
- |
PREDICTED: uncharacterized protein LOC103939917 [Pyrus x bretschneideri] |
| 4 |
Hb_000163_240 |
0.1771310539 |
- |
- |
PREDICTED: protein OPI10 homolog [Jatropha curcas] |
| 5 |
Hb_008406_040 |
0.1779590764 |
- |
- |
- |
| 6 |
Hb_000218_020 |
0.1801845998 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 7 |
Hb_001454_220 |
0.1819272523 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 8 |
Hb_002450_140 |
0.1819727443 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 9 |
Hb_000116_510 |
0.1852648542 |
- |
- |
PREDICTED: RNA-binding protein 24-A-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_013394_060 |
0.1859195702 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 11 |
Hb_003228_030 |
0.1934836462 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_000488_010 |
0.1950836694 |
- |
- |
18.1 kDa class I heat shock protein [Jatropha curcas] |
| 13 |
Hb_002400_230 |
0.1952301314 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 14 |
Hb_003026_030 |
0.1955622055 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 15 |
Hb_002311_140 |
0.1957925129 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 16 |
Hb_003018_040 |
0.1969724376 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000997_150 |
0.1981261252 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_031711_010 |
0.1981999658 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001277_360 |
0.1991300461 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 20 |
Hb_002999_060 |
0.2008089475 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |