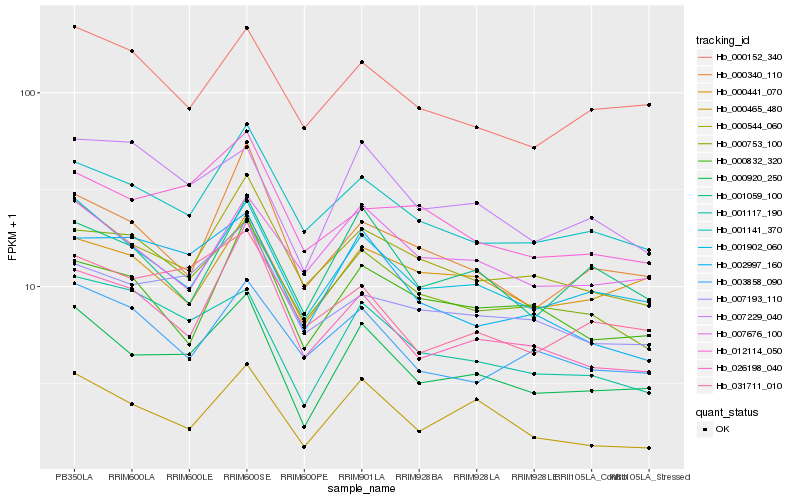
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_250 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein RCOM_0655500 [Ricinus communis] |
| 2 |
Hb_001902_060 |
0.0770198116 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000544_060 |
0.0996012113 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 4 |
Hb_000465_480 |
0.1035047815 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16835, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001141_370 |
0.1061907517 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 6 |
Hb_001117_190 |
0.1093868868 |
transcription factor |
TF Family: FAR1 |
FAR1-related sequence 11 [Theobroma cacao] |
| 7 |
Hb_001059_100 |
0.1153941193 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein U-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000753_100 |
0.1156704147 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007229_040 |
0.1160426435 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 10 |
Hb_000152_340 |
0.1163976415 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 11 |
Hb_026198_040 |
0.1175715296 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 12 |
Hb_007193_110 |
0.1187127929 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 13 |
Hb_031711_010 |
0.1203594439 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000340_110 |
0.1225521105 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 15 |
Hb_002997_160 |
0.1227893304 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000441_070 |
0.1228304875 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007676_100 |
0.1232490975 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 18 |
Hb_012114_050 |
0.1240820037 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000832_320 |
0.1253285502 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_003858_090 |
0.126127438 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |