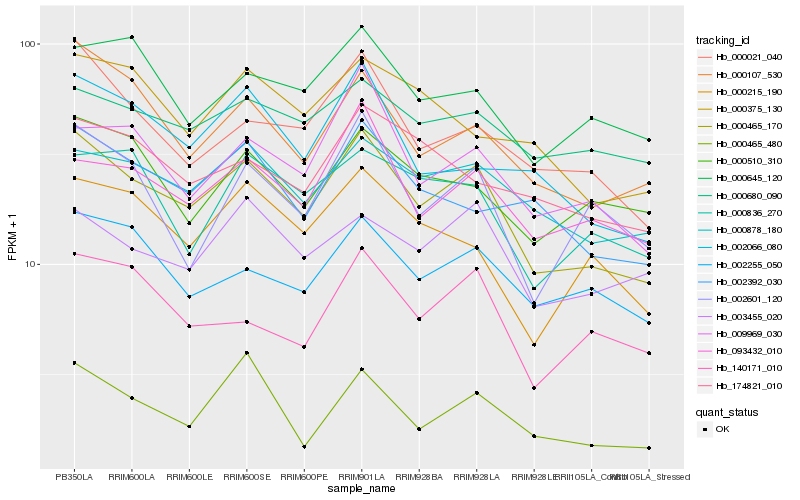
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_170 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 2 |
Hb_000107_530 |
0.0934447465 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000878_180 |
0.098343996 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000021_040 |
0.099005166 |
- |
- |
PREDICTED: villin-3-like isoform X1 [Populus euphratica] |
| 5 |
Hb_093432_010 |
0.0999125664 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 6 |
Hb_002255_050 |
0.1045936307 |
- |
- |
PREDICTED: probable isoleucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 7 |
Hb_000215_190 |
0.1069271012 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |
| 8 |
Hb_009969_030 |
0.1071076571 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002392_030 |
0.1072545152 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 10 |
Hb_002066_080 |
0.1080939575 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 11 |
Hb_003455_020 |
0.1107574711 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 12 |
Hb_140171_010 |
0.1118806999 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 13 |
Hb_002601_120 |
0.1130207479 |
- |
- |
PREDICTED: uncharacterized protein LOC105634921 [Jatropha curcas] |
| 14 |
Hb_000645_120 |
0.1136414943 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 15 |
Hb_000510_310 |
0.1149659315 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 16 |
Hb_000836_270 |
0.115536849 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 17 |
Hb_000680_090 |
0.1155743908 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 18 |
Hb_174821_010 |
0.1167145247 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 19 |
Hb_000375_130 |
0.1168891313 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000465_480 |
0.1175265525 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16835, mitochondrial [Jatropha curcas] |