| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
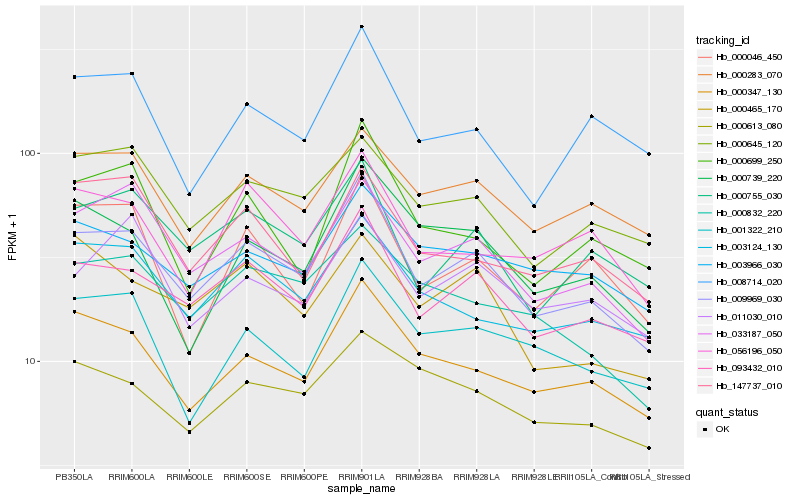
Hb_009969_030 |
0.0 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_093432_010 |
0.0542428773 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 3 |
Hb_033187_050 |
0.0812000407 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 4 |
Hb_000347_130 |
0.0820407442 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 5 |
Hb_000755_030 |
0.082783801 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 6 |
Hb_003124_130 |
0.0916180352 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000283_070 |
0.0934172426 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 8 |
Hb_000645_120 |
0.0956436632 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 9 |
Hb_003966_030 |
0.0974219534 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X7 [Jatropha curcas] |
| 10 |
Hb_056196_050 |
0.0985076834 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 11 |
Hb_000832_220 |
0.0991168823 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X1 [Jatropha curcas] |
| 12 |
Hb_008714_020 |
0.0994282022 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 13 |
Hb_000699_250 |
0.1025703679 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000739_220 |
0.1036645851 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 15 |
Hb_000613_080 |
0.1053382195 |
- |
- |
PREDICTED: mucin-5AC-like [Nicotiana sylvestris] |
| 16 |
Hb_001322_210 |
0.1057357146 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6-like [Jatropha curcas] |
| 17 |
Hb_147737_010 |
0.1061083986 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000046_450 |
0.1068656427 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 19 |
Hb_000465_170 |
0.1071076571 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 20 |
Hb_011030_010 |
0.1078865482 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |