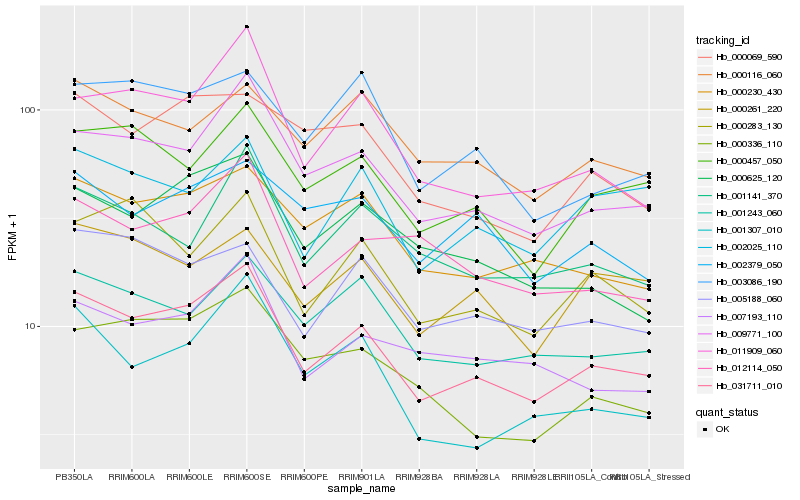
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031711_010 |
0.0 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_009771_100 |
0.065114114 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 3 |
Hb_011909_060 |
0.0870026025 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 4 |
Hb_000230_430 |
0.0876429259 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_000457_050 |
0.0882882612 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 6 |
Hb_001243_060 |
0.0934051471 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 7 |
Hb_007193_110 |
0.0954887641 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 8 |
Hb_000625_120 |
0.0960535281 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 9 |
Hb_000283_130 |
0.0970312095 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 10 |
Hb_005188_060 |
0.0997430208 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 11 |
Hb_000336_110 |
0.0997548231 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_001141_370 |
0.1008134187 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 13 |
Hb_012114_050 |
0.1017222422 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000069_590 |
0.1028758816 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 15 |
Hb_001307_010 |
0.1030820241 |
- |
- |
PREDICTED: uncharacterized protein LOC103939917 [Pyrus x bretschneideri] |
| 16 |
Hb_000261_220 |
0.1045228371 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 17 |
Hb_003086_190 |
0.1049725709 |
- |
- |
PREDICTED: uncharacterized protein LOC105635794 [Jatropha curcas] |
| 18 |
Hb_002379_050 |
0.1058250705 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 19 |
Hb_000116_060 |
0.1060155438 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 20 |
Hb_002025_110 |
0.1061399631 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |