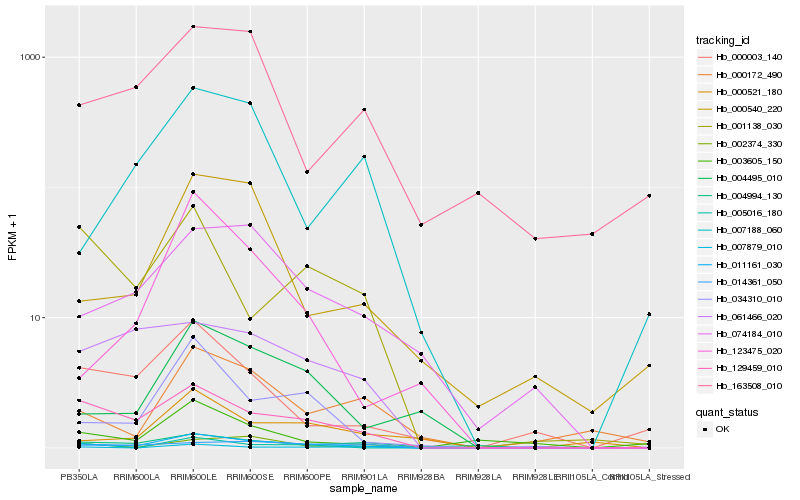
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011161_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638906 [Jatropha curcas] |
| 2 |
Hb_129459_010 |
0.1651923169 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 3 |
Hb_034310_010 |
0.2005013318 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_004495_010 |
0.2117444327 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 5 |
Hb_074184_010 |
0.2264536147 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 6 |
Hb_002374_330 |
0.2408019804 |
- |
- |
- |
| 7 |
Hb_000172_490 |
0.2408912817 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 8 |
Hb_007879_010 |
0.2413073753 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 9 |
Hb_061466_020 |
0.2449513954 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 10 |
Hb_004994_130 |
0.247893549 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |
| 11 |
Hb_014361_050 |
0.2492822592 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000521_180 |
0.2493494736 |
- |
- |
PREDICTED: uncharacterized protein LOC105650335 [Jatropha curcas] |
| 13 |
Hb_123475_020 |
0.2525469454 |
- |
- |
- |
| 14 |
Hb_000540_220 |
0.2530883262 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 15 |
Hb_000003_140 |
0.2560776747 |
- |
- |
hypothetical protein glysoja_047034, partial [Glycine soja] |
| 16 |
Hb_007188_060 |
0.2632946328 |
- |
- |
- |
| 17 |
Hb_001138_030 |
0.2656137343 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 18 |
Hb_005016_180 |
0.2657314051 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 19 |
Hb_003605_150 |
0.2708035427 |
- |
- |
chitinase family protein [Populus trichocarpa] |
| 20 |
Hb_163508_010 |
0.2714701222 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |