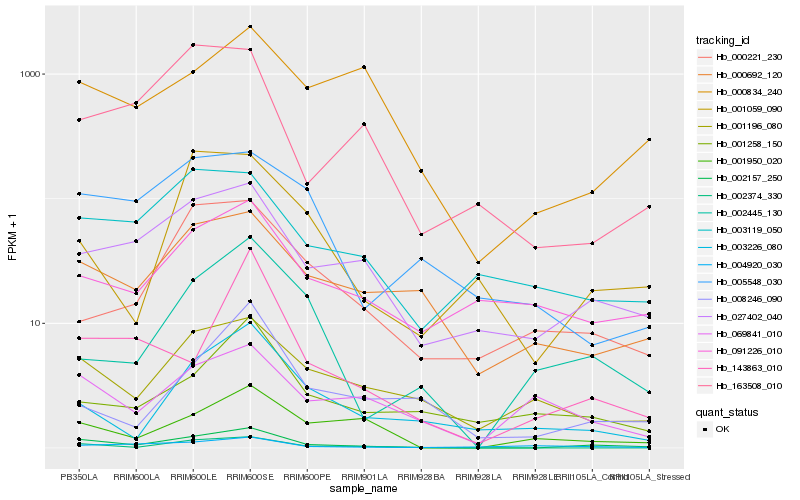
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002157_250 |
0.0 |
- |
- |
Pectinesterase-4 precursor, putative [Ricinus communis] |
| 2 |
Hb_027402_040 |
0.2146901637 |
- |
- |
Thioredoxin II, putative [Ricinus communis] |
| 3 |
Hb_002374_330 |
0.2161013467 |
- |
- |
- |
| 4 |
Hb_001196_080 |
0.2408253207 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 5 |
Hb_000692_120 |
0.2418536657 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 6 |
Hb_001950_020 |
0.250831084 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 7 |
Hb_002445_130 |
0.255945162 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 8 |
Hb_003119_050 |
0.2562770463 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001059_090 |
0.2568738268 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |
| 10 |
Hb_003226_080 |
0.2580298599 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |
| 11 |
Hb_143863_010 |
0.2607217096 |
- |
- |
PREDICTED: uncharacterized protein LOC105113187 isoform X2 [Populus euphratica] |
| 12 |
Hb_069841_010 |
0.2631413502 |
- |
- |
resistance protein [Quercus suber] |
| 13 |
Hb_163508_010 |
0.263225329 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 14 |
Hb_091226_010 |
0.2651000724 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 15 |
Hb_000834_240 |
0.2661130434 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 16 |
Hb_000221_230 |
0.2666863767 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 17 |
Hb_005548_030 |
0.2671287364 |
- |
- |
hypothetical protein JCGZ_05606 [Jatropha curcas] |
| 18 |
Hb_004920_030 |
0.2673743355 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase CKI1 [Jatropha curcas] |
| 19 |
Hb_008246_090 |
0.2679713309 |
- |
- |
- |
| 20 |
Hb_001258_150 |
0.2680550798 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |