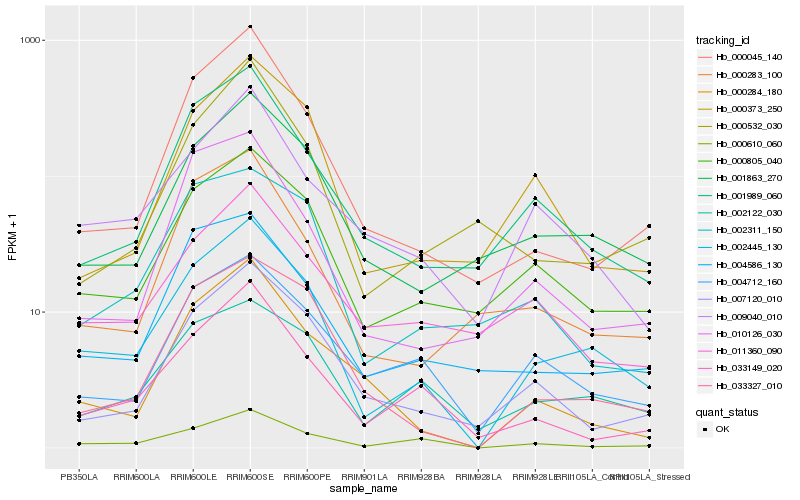
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002445_130 |
0.0 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 2 |
Hb_000805_040 |
0.1425368759 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 3 |
Hb_001863_270 |
0.1481812227 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 4 |
Hb_000610_060 |
0.1530343826 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 5 |
Hb_001989_060 |
0.1597541687 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 6 |
Hb_033327_010 |
0.160810286 |
- |
- |
SYM8 [Pisum sativum] |
| 7 |
Hb_002122_030 |
0.1610489487 |
- |
- |
- |
| 8 |
Hb_009040_010 |
0.1635549241 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000045_140 |
0.1666952652 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 10 |
Hb_004712_160 |
0.1678241517 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 11 |
Hb_004586_130 |
0.1678603145 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007120_010 |
0.171341232 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 13 |
Hb_033149_020 |
0.1720138832 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 14 |
Hb_000373_250 |
0.1741805299 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 15 |
Hb_000283_100 |
0.176827012 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000284_180 |
0.1816554605 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 17 |
Hb_010126_030 |
0.1821311005 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011360_090 |
0.1837338909 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 19 |
Hb_002311_150 |
0.1851514504 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 20 |
Hb_000532_030 |
0.1857478802 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |