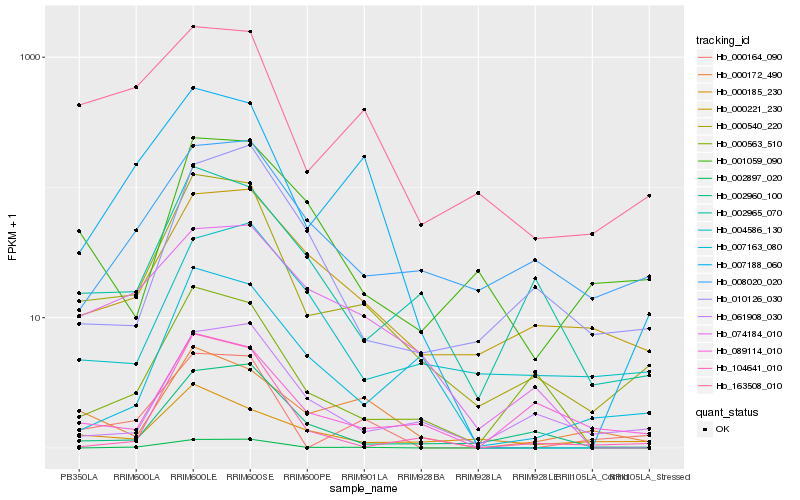
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000540_220 |
0.0 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 2 |
Hb_163508_010 |
0.1751270526 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 3 |
Hb_002965_070 |
0.1888726808 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 4 |
Hb_000164_090 |
0.1904914359 |
- |
- |
hypothetical protein [Plasmodium berghei strain ANKA], partial [Plasmodium berghei ANKA] |
| 5 |
Hb_007188_060 |
0.1915386523 |
- |
- |
- |
| 6 |
Hb_000221_230 |
0.192203041 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 7 |
Hb_000563_510 |
0.1961485391 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 8 |
Hb_002960_100 |
0.1963650012 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 9 |
Hb_074184_010 |
0.200173383 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 10 |
Hb_061908_030 |
0.2042364389 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 11 |
Hb_089114_010 |
0.2100389934 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 12 |
Hb_002897_020 |
0.2138648474 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 13 |
Hb_008020_020 |
0.2159835391 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_004586_130 |
0.2168105971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000172_490 |
0.2183998027 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 16 |
Hb_007163_080 |
0.2189389734 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 17 |
Hb_010126_030 |
0.2190592444 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000185_230 |
0.2193389962 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_104641_010 |
0.2218045373 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001059_090 |
0.2272730647 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |