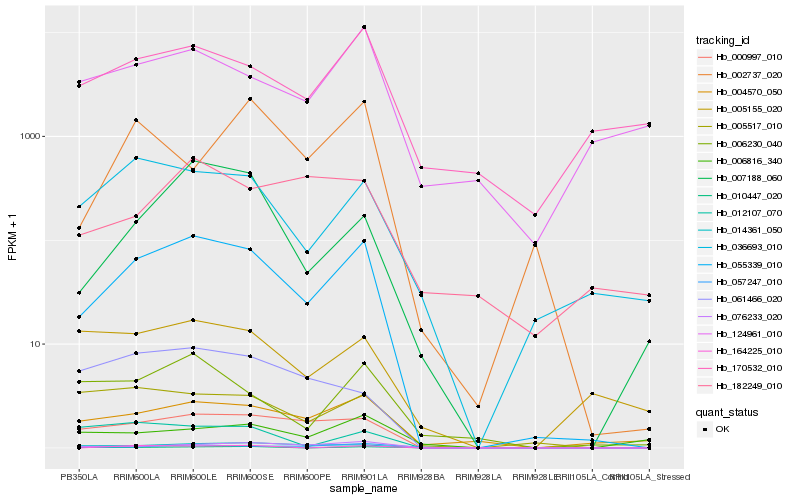
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055339_010 |
0.0 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 2 |
Hb_000997_010 |
0.1793060946 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 3 |
Hb_076233_020 |
0.2047403649 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Populus euphratica] |
| 4 |
Hb_014361_050 |
0.2064959522 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004570_050 |
0.2121852211 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_036693_010 |
0.2218697719 |
- |
- |
- |
| 7 |
Hb_007188_060 |
0.2227532387 |
- |
- |
- |
| 8 |
Hb_005517_010 |
0.2357227918 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_006230_040 |
0.2366200892 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_170532_010 |
0.2496931225 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 11 |
Hb_057247_010 |
0.2528328714 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 12 |
Hb_061466_020 |
0.2556411027 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 13 |
Hb_124961_010 |
0.2592283113 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 14 |
Hb_164225_010 |
0.2635575543 |
- |
- |
- |
| 15 |
Hb_005155_020 |
0.2639071504 |
- |
- |
- |
| 16 |
Hb_012107_070 |
0.2656342649 |
- |
- |
- |
| 17 |
Hb_010447_020 |
0.2699313937 |
- |
- |
PREDICTED: uncharacterized protein LOC105797464 [Gossypium raimondii] |
| 18 |
Hb_002737_020 |
0.2715220247 |
- |
- |
metallothionein [Bruguiera gymnorhiza] |
| 19 |
Hb_006816_340 |
0.2720727688 |
- |
- |
- |
| 20 |
Hb_182249_010 |
0.2725498276 |
- |
- |
- |