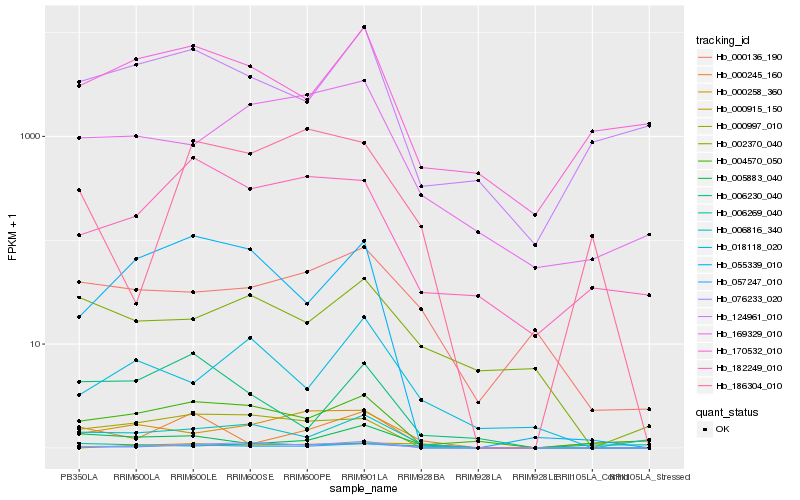
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_057247_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 2 |
Hb_005883_040 |
0.2154043638 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 3 |
Hb_076233_020 |
0.2159405227 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Populus euphratica] |
| 4 |
Hb_004570_050 |
0.2303237523 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_006816_340 |
0.2346786681 |
- |
- |
- |
| 6 |
Hb_000136_190 |
0.2401139205 |
- |
- |
PREDICTED: protein-S-isoprenylcysteine O-methyltransferase A-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_182249_010 |
0.2439400234 |
- |
- |
- |
| 8 |
Hb_000997_010 |
0.2493171187 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 9 |
Hb_055339_010 |
0.2528328714 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 10 |
Hb_169329_010 |
0.2538120634 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000258_360 |
0.2555499529 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 12 |
Hb_124961_010 |
0.2594245648 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 13 |
Hb_170532_010 |
0.2615484078 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 14 |
Hb_002370_040 |
0.2688699357 |
- |
- |
PREDICTED: uncharacterized protein LOC105644336 [Jatropha curcas] |
| 15 |
Hb_000915_150 |
0.2694551483 |
- |
- |
- |
| 16 |
Hb_018118_020 |
0.2704132036 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_006269_040 |
0.272045853 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 18 |
Hb_186304_010 |
0.273081575 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 19 |
Hb_006230_040 |
0.2736254846 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_000245_160 |
0.2742444 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |