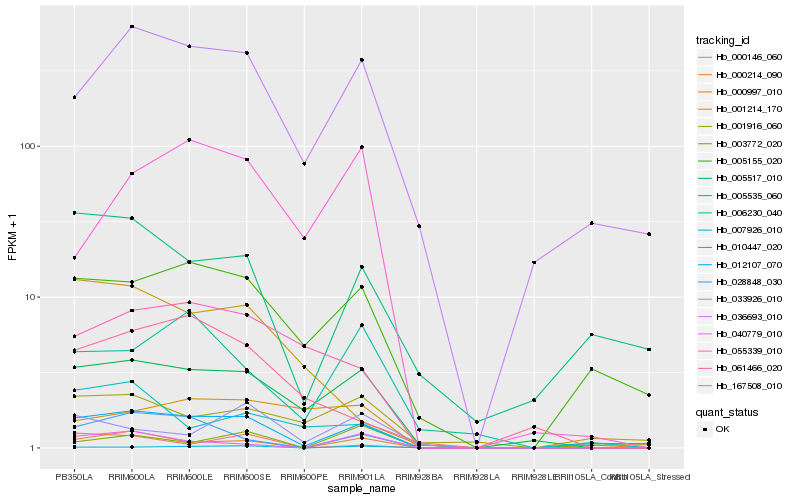
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012107_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_005517_010 |
0.1564526474 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_036693_010 |
0.2015084756 |
- |
- |
- |
| 4 |
Hb_028848_030 |
0.2104567583 |
- |
- |
- |
| 5 |
Hb_005155_020 |
0.2229475415 |
- |
- |
- |
| 6 |
Hb_061466_020 |
0.2316780892 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 7 |
Hb_000214_090 |
0.2330533078 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 8 |
Hb_040779_010 |
0.2370719206 |
- |
- |
- |
| 9 |
Hb_006230_040 |
0.2385089165 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_005535_060 |
0.2516086177 |
- |
- |
- |
| 11 |
Hb_000146_060 |
0.2535942453 |
- |
- |
- |
| 12 |
Hb_001916_060 |
0.2576818385 |
- |
- |
PREDICTED: putative protein FAR1-RELATED SEQUENCE 10 isoform X2 [Jatropha curcas] |
| 13 |
Hb_010447_020 |
0.2580675972 |
- |
- |
PREDICTED: uncharacterized protein LOC105797464 [Gossypium raimondii] |
| 14 |
Hb_055339_010 |
0.2656342649 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 15 |
Hb_167508_010 |
0.2662129522 |
- |
- |
hypothetical protein POPTR_0001s27330g [Populus trichocarpa] |
| 16 |
Hb_033926_010 |
0.2714080953 |
- |
- |
- |
| 17 |
Hb_007926_010 |
0.2756832858 |
- |
- |
PREDICTED: UPF0554 protein C2orf43 homolog isoform X4 [Citrus sinensis] |
| 18 |
Hb_001214_170 |
0.2806285634 |
- |
- |
pyruvate kinase family protein [Medicago truncatula] |
| 19 |
Hb_000997_010 |
0.2821884727 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003772_020 |
0.2870892585 |
- |
- |
PREDICTED: uncharacterized protein LOC105037131, partial [Elaeis guineensis] |