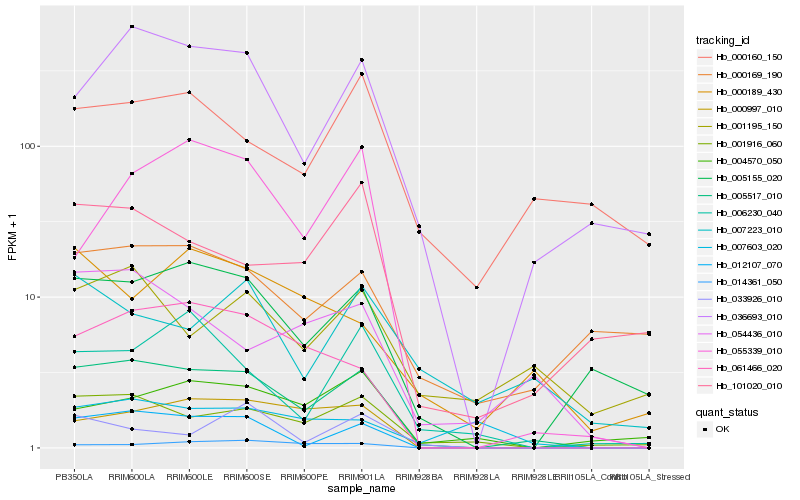
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005517_010 |
0.0 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_012107_070 |
0.1564526474 |
- |
- |
- |
| 3 |
Hb_036693_010 |
0.177650786 |
- |
- |
- |
| 4 |
Hb_001916_060 |
0.1829031795 |
- |
- |
PREDICTED: putative protein FAR1-RELATED SEQUENCE 10 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005155_020 |
0.1909463071 |
- |
- |
- |
| 6 |
Hb_061466_020 |
0.1946440126 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 7 |
Hb_000997_010 |
0.2028811516 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 8 |
Hb_054436_010 |
0.2075606217 |
- |
- |
PREDICTED: uncharacterized protein LOC105638666 [Jatropha curcas] |
| 9 |
Hb_006230_040 |
0.2250040687 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_014361_050 |
0.2254000898 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 11 |
Hb_101020_010 |
0.2318158141 |
- |
- |
- |
| 12 |
Hb_001195_150 |
0.2341651243 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 13 |
Hb_055339_010 |
0.2357227918 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 14 |
Hb_000189_430 |
0.241405879 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000169_190 |
0.241425613 |
desease resistance |
Gene Name: AAA_17 |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 16 |
Hb_000160_150 |
0.2440982256 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Cucumis melo] |
| 17 |
Hb_004570_050 |
0.2448442854 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_007223_010 |
0.249319276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007603_020 |
0.251347017 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 20 |
Hb_033926_010 |
0.2514366094 |
- |
- |
- |