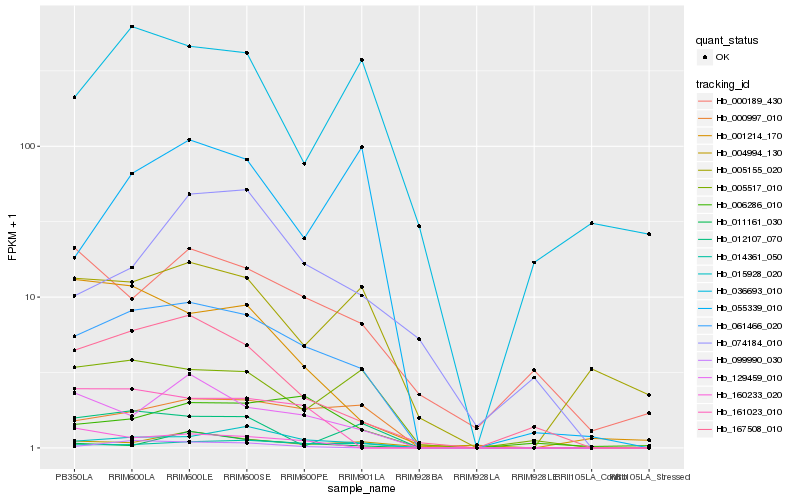
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061466_020 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 2 |
Hb_015928_020 |
0.1632067864 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 3 |
Hb_000997_010 |
0.1687710605 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 4 |
Hb_014361_050 |
0.1734707728 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 5 |
Hb_167508_010 |
0.1791999604 |
- |
- |
hypothetical protein POPTR_0001s27330g [Populus trichocarpa] |
| 6 |
Hb_129459_010 |
0.1799224649 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 7 |
Hb_005517_010 |
0.1946440126 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_006286_010 |
0.2273301722 |
- |
- |
hypothetical protein PRUPE_ppa017148mg [Prunus persica] |
| 9 |
Hb_161023_010 |
0.2285140941 |
- |
- |
PREDICTED: uncharacterized protein LOC105639929 [Jatropha curcas] |
| 10 |
Hb_005155_020 |
0.2316240391 |
- |
- |
- |
| 11 |
Hb_012107_070 |
0.2316780892 |
- |
- |
- |
| 12 |
Hb_000189_430 |
0.2364026653 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_001214_170 |
0.2378406243 |
- |
- |
pyruvate kinase family protein [Medicago truncatula] |
| 14 |
Hb_011161_030 |
0.2449513954 |
- |
- |
PREDICTED: uncharacterized protein LOC105638906 [Jatropha curcas] |
| 15 |
Hb_036693_010 |
0.2451639626 |
- |
- |
- |
| 16 |
Hb_099990_030 |
0.2525908064 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 17 |
Hb_074184_010 |
0.2539628234 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 18 |
Hb_055339_010 |
0.2556411027 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 19 |
Hb_004994_130 |
0.2585155832 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |
| 20 |
Hb_160233_020 |
0.2656631271 |
- |
- |
- |